Vol 100, No 2 (2023)
- Year: 2023
- Published: 22.05.2023
- Articles: 12
- URL: https://microbiol.crie.ru/jour/issue/view/55
ORIGINAL RESEARCHES
Virus-inhibitory activity of the antigen complex of opportunistic pathogenic bacteria against SARS-CoV-2
Abstract
Introduction. The antigen complex of opportunistic pathogenic bacteria (ACOPB) has a protective effect against avian influenza viruses, herpes virus type 2, and other viruses that cause acute respiratory viral infections. In the context of the COVID-19 pandemic, an important task is to find out whether ACOPB has a protective effect against SARS-CoV-2.
The purpose of the study was to evaluate in vitro the ACOPB virus-inhibitory activity against the Dubrovka laboratory strain of SARS-CoV-2.
Materials and methods. The study was performed using Vero cell line CCL-81, human peripheral blood mononuclear cells (PBMCs), mouse monoclonal anti-idiotypic antibodies structurally mimicking biological effects of human interferons (IFNs), the Dubrovka laboratory strain of SARS-CoV-2. The infectivity of the virus was assessed by two methods: by virus titration using cell cultures and the limiting dilution method when the results are assessed by a cytopathic effect; the second method was a plaque assay. The in vitro virus inhibition test was performed using the cell culture susceptible to SARS-CoV-2; the mixture containing a specific dose of the virus and a two-fold dilution of ACOPB was transferred to the cell culture after the ACOPB medication had interacted with the virus at 4ºC for 2 hours. The ACOPB virus-inhibitory activity against SARS-CoV-2 was assessed by the functional activity of α/β and γ IFN receptors (RIFN) in human PBMCs induced in vitro by ACOPB and the ACOPB mixture with the specific dose of SARS-CoV-2. The RIFN expression level was measured by the indirect membrane immunofluorescence test.
Results. Hemagglutination assay using chicken, mouse, guinea pig, and human red blood cells was performed for detection of the SARS-CoV-2 inhibitory protein. The lysate of Vero CCL-81 cells infected with SARS-CoV-2 Dubrovka demonstrated the highest hemagglutination activity with guinea pig red blood cells and low titers of hemagglutination in the virus-containing fluid. The virus inhibition test in the Vero CCL-81 cell culture demonstrated that ACOPB inhibited 10 doses of SARS-CoV-2 Dubrovka with the titer 1 : 32, providing 100% protection of the cell culture for 8 days (the monitoring period). ACOPB induced α/β and γ RIFN expression on membranes of human PBMCs in in vitro cultures and decreased RIFN α/β and γ expression after its interaction with SARS-CoV-2 Dubrovka.
Conclusion. The experimental studies including the virus inhibition test in the cell culture susceptible to SARS-CoV-2 Dubrovka and the indirect membrane immunofluorescence assay using monoclonal anti-idiotypic antibodies mimicking IFN-like properties demonstrated that ACOPB had both an immunomodulatory and a virus-inhibitory effect.
 143-152
143-152


The structure of ESKAPE pathogens isolated from patients of the neonatal intensive care unit at the National Hospital of Pediatrics in Hanoi, the Socialist Republic of Vietnam
Abstract
Introduction. The incidence of healthcare-associated infections is a major public health problem worldwide, affecting all countries regardless of their economic status. The main agents of these infections are pathogens belonging to the ESKAPE group.
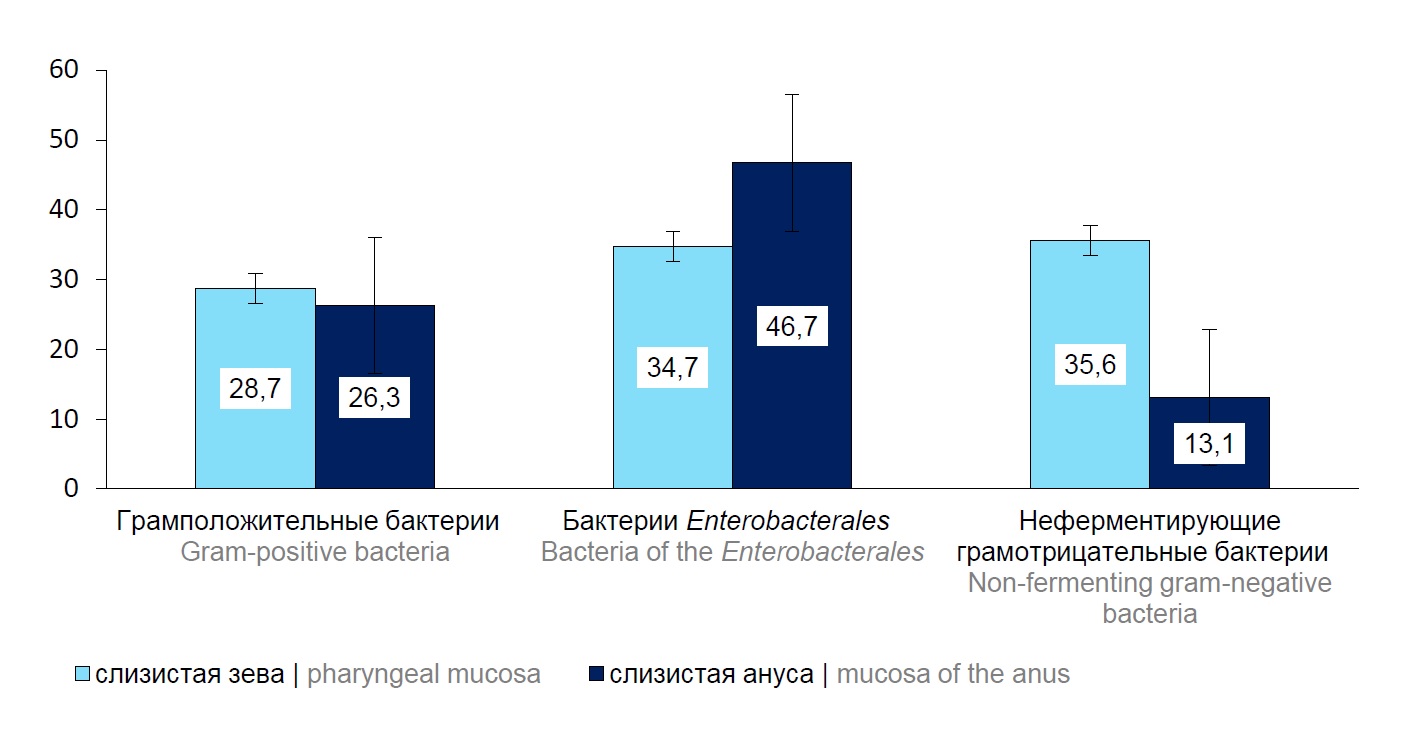
The aim of the study was to explore the structure, molecular and antigenic characteristics of the ESKAPE pathogens isolated from oral and anal mucosa of patients of the neonatal intensive care unit (NICU), and to assess their etiological significance in occurrence of healthcare-associated infections.
Materials and methods. Samples from a total of 49 children were tested, including 40 newborns – patients of NICU at the National Hospital of Pediatrics in Hanoi. Collection and processing of biomaterial (oropharyngeal swabs, rectal swabs) and isolation of bacterial cultures were performed using conventional bacteriological methods. Mass spectrometry was used for identification of isolates. Klebsiella pneumoniae strains were analyzed using the whole-genome sequencing method.
Results. The group of gram-positive ESKAPE pathogens identified in oral mucosa was represented by isolates Enterococcus faecium and Staphylococcus aureus. The isolates of the family Enterobacteriaceae included K. pneumoniae, Escherichia coli, Enterobacter cloacae; the group of nonfermenting gram-negative bacteria was represented by Pseudomonas aeruginosa, Acinetobacter baumannii. The structure of ESKAPE pathogens persistent in anal mucosa was characterized by dominance of Enterococcus spp., E. coli, K. pneumoniae and P. aeruginosa bacteria. The whole-genome sequencing of K. pneumoniae isolates revealed 7 clusters and 8 sequence types. ST14 and ST1741 prevailed, accounting for 25%, respectively, of the total number of the studied strains. The molecular serotyping showed that by the O antigen, strains belonged mainly to serotypes O1v1, O1/ O2v2, O5; by the presence of the capsular antigen — to serotypes KL2, KL104, KL60.
Conclusion. The analysis of the structure of the ESKAPE pathogens isolated from the oral and anal mucosa of patients of NICU at the National Hospital of Pediatrics in Hanoi identified etiologically significant agents of bacterial infections: S. aureus, K. pneumoniae, E. coli, E. cloacae, P. aeruginosa, A. baumannii. The molecular and genetic analysis of K. pneumoniae strains co-circulating in mucous membranes of several patients of the unit revealed their homology, thus confirming healthcare-associated contamination of children with nosocomial strains.
 168-177
168-177


Development of a method for molecular subtyping Bacillus anthracis using HRM PCR
Abstract
Introduction. Bacillus anthracis is the causative agent of anthrax, a pathogen characterized by high genetic monomorphism that complicates differentiation of strains. Thus, molecular methods for pathogen typing require the improvement.
The aim of the study. To select marker SNPs for new genetic groups of B. anthracis and to develop a method for their laboratory identification using HRM PCR.
Materials and methods. The core genome of 222 strains of B. anthracis from the GenBank database and 66 strains from the collection of pathogenic microorganisms of the Stavropol Anti-Plague Institute was aligned using the parsnp software. A dendrogram based on 7242 core genome SNPs was built in MEGA X software. The strains for validation of the HRM method included representatives of various genetic groups. The HRM PCR reaction was performed using the "Type-it HRM PCR Kit" and "KAPA HRM FAST qPCR Kit" and a Rotor Gene DNA thermocycler with HRM function. Data analysis and visualization were performed using custom scripts in the Python and R development environments.
Results and discussion. Marker SNPs for 6 genetic groups have been identified, which make it possible to determine whether strains belong to one of 7 new subclusters. Pairs of primers were selected for the loci containing them, HRM PCR parameters were optimized for discrimination of different alleles of SNP loci, and an analysis scheme was developed.
Conclusion. Thus, marker SNPs were selected to determine the genetic subclusters A.Br.CEA, A.Br.STI, A.Br.Tsiankovskii, B.Br.Europe, B.Br.Siberia, B.Br.Asia, B.Br.018, and a new laboratory method was developed for molecular subtyping of B. anthracis using HRM PCR.
 178-187
178-187


Humoral immunity against pertussis among healthcare workers
Abstract
Background. Vaccination is the most effective way to prevent infectious diseases. Inadequate vaccination coverage among healthcare workers is a major concern for healthcare organizations. The lack of specific immunity against pertussis represents the risk for acquiring a healthcare associated infection by medical staff but also of being a source of infection to patients. More than 70% of all healthcare associated infections are vaccine-preventable. Pertussis remains one of the most important vaccine-preventable infections and continues to be a public health concern even in countries with high vaccination coverage among children. Revaccination of adults against pertussis is not included in the National vaccination schedule of the Russian Federation.
Aim. To assess the humoral immunity against pertussis among healthcare workers of the infectious disease hospitals.
Materials and methods. We conducted a cross-sectional study that included 252 healthcare workers. All study participants were surveyed and tested for antibodies (immunoglobulins G) against Bordetella pertussis by enzyme immunoassay.
Results. The proportion of healthcare workers seronegative for pertussis was 46.8%. The healthcare workers with unknown vaccination status amounted to 40.5%. More than half (55.6%) of the participants have been vaccinated and 3.9% of them have had pertussis in childhood. A recent infection was confirmed in 8.0% of participants who had the level of antibodies to Bordetella pertussis greater than 50 U/ml. The largest proportion of participants seronegative to Bordetella pertussis (55.2%) was observed among those under 30 years. The median level of antibodies against pertussis in seropositive health workers was 28.3 U/ml.
Conclusion. The significant proportion of seronegative participants (46.8%) and those who had the recent infection underline the necessity of the improvement of pertussis prevention by implementation of vaccination in the risk groups, including healthcare workers to reduce the risk of healthcare associated infections.
 196-202
196-202


Antimicrobial resistance in foodborne Salmonella enterica isolates in the Republic of Belarus
Abstract
Introduction. Antimicrobial resistance is a global public health concern. Salmonella spp., which can be transmitted to humans through contaminated food, are among the most important foodborne pathogens worldwide.
Materials and methods. The antimicrobial resistance of 358 bacterial isolates collected from food and water in the Republic of Belarus (Belarus) in 2018–2021 was studied by analyzing phenotypic and genotypic characteristics of antibiotic bacterial resistance. MALDI-TOF mass spectrometry was used to classify and identify bacteria. Phenotypic antimicrobial susceptibility of bacteria was measured by the minimum inhibitory concentration method using a Sensititre automated bacteriological analyzer and the disk diffusion test for 45 antimicrobial agents. Antimicrobial resistance genes in multidrug-resistant Salmonella isolates were identified by whole-genome sequencing.
Results. The in vitro testing of phenotypic bacterial susceptibility showed high susceptibility to fluoroquinolones (97.2%), third-generation cephalosporins (93.9%), carbapenems (98.0%), ampicillin (81.8%), aminoglycosides (97.5%), tetracyclines (87.5%), chloramphenicol (93.8%), trimethoprim/sulfamethoxazole (co-trimoxazole) (95.3%) and colistin (85.2%). It was found that the antibiotic resistance mechanism in S. enterica was associated with the presence of genes blaTEM-1B (82%), blaTEM-1C (7.7%), blaSHV-12 (2.6%), blaDHA-1 (2.6%), blaCMY-2 (7.7%), qnrB2 (9.1%), qnrB4 (9.1%), qnrB5 (9.1%), qnrB19 (72.7%), aac(6’)-Ib-cr (9.1%), aac(6’)-Iaa (100%), aadA1 (13.2%), aadA2 (8.8%), tetB (74.3%), tetA (25.7%), tetM (2.9%), tetD (28.6%), mcr-9 (1.5%).
Conclusion. All the bacterial isolates were phenotypically susceptible to first-line antibiotics used in treatment of salmonellosis: fluoroquinolones and third-generation cephalosporins. The whole-genome sequencing of multidrug-resistant Salmonella isolates (19.0%) detected resistance genes for 9 groups of antibiotics: aminoglycosides (100%), beta-lactams (57.4%), fluoroquinolones (16.2%), tetracyclines (51.5%), macrolides (1.5%), phenicols (30.4%), trimethoprim (13.0%), sulfonamides (47.8%) and colistin (1.4%). Thus, epidemiological surveillance of the Salmonella spread through the food chain is of critical importance for the monitoring of antimicrobial resistance among foodborne Salmonella.
 153-167
153-167


Antibiotic sensitivity of cholera vibrions in biofilms formed on various substrates
Abstract
Introduction. Evaluation of antibiotic sensitivity of biofilms formed by Vibrio cholerae can help in the search for effective drugs to combat cholera.
The aim of the work is to evaluate the effectiveness of antibacterial drugs against V. cholerae cells being a part of a polymicrobial biofilm formed on various substrates.
Materials and methods. Mono- and polymicrobial biofilms were simulated in vitro on chitin and plastic in flasks containing tap autoclaved water contaminated with a suspension of 104 microbial cells of three strains of V. cholerae O1 El Tor separately or in combination with Escherichia coli QD 5003.
Results. When being a part of mono- or polymicrobial bacterial communities formed by the studied strains on chitin or plastic, the V. cholerae and E. coli cells were less susceptible to the action of antibacterial drugs.
Conclusion. The increased antibiotic resistance of V. cholerae biofilms formed on biotic and abiotic surfaces highlights the danger of their spread and preservation in the environment, creates additional problems regarding the use of antibiotics and requires the development of alternative strategies to reduce resistance.
 188-195
188-195


Antigenic identity of immunodominant proteins of Bacillus anthracis genovariants
Abstract
Introduction. The main biological raw materials for the production of immunobiological preparations for identification of Bacillus anthracis are its specific antigens, the protective antigen and the EA1 protein.
Purpose. To determine the antigenic identity of immunodominant proteins of different genovariants of B. anthracis isolated by gel chromatography and electrophoresis.
Materials and methods. Culture filtrates of isogenic variants of B. anthracis strain 575/122 (pXO1+, pXO2+): R01 (pXO1+, pXO2–); R00 (pXO1–, pXO2–) were used in the study. Gel chromatographic fractionation and electrophoretic separation were carried out according to standard methods. The antigenic properties of proteins isolated by gel chromatography and electrophoresis were studied by immunodiffusion with polyclonal monospecific sera against the protective antigen and the EA1 protein of the S-layer.
Results. Gel chromatographic separation of B. anthracis 575/122 culture filtrates R01 (pXO1+, pXO2–) and R00 (pXO1–, pXO2–)yielded fractions 1 and 5. Sera against EA1 protein and antigens of fraction 1 of strains B. anthracis 575/122 R00 and B. anthracis 575/122 R01 culture filtrates identified the identical antigens. Serum against antigens of fraction 5 of B. anthracis 575/122 R01 contained antibodies to numerous proteins, including the protective antigen isolated by electrophoresis.
Discussion. The antigenic identity of immunodominant proteins isolated by gel chromatography and electrophoresis was identified.
Conclusion. EA1 and PA proteins isolated by electrophoresis and gel chromatography can be used for production of monoclonal and polyclonal monospecific antibodies suitable for the design of diagnostic preparations.
 203-208
203-208


Influence of povidone-yodine on the sensitivity of clinical isolates of Klebsiella pneumoniae to antibiotics
Abstract
Introduction. Cross-resistance of microorganisms to antibiotics against the background of the use of biocides in subinhibitory concentrations is an urgent problem of modern health care.
The aim of the work is to study the effect of povidone-iodine on the sensitivity of clinical isolates of Klebsiella pneumoniae to antibiotics.
Materials and methods. The work analyzed the effect of povidone-iodine at subinhibitory concentrations on changes in the sensitivity of clinical isolates of K. pneumoniae (n = 9) to antibacterial agents (n = 15). Adaptation of bacteria to povidone-iodine was carried out using periodic cultivation of microorganisms, which was carried out in 96-well culture plates for suspension cultures (non-treated) without stirring. The sensitivity of bacteria to antibiotics was assessed using an automatic analyzer “Vitek2Compact” (“BioMerieux”) using AST-204 charts.
Results. During the adaptation of clinical isolates of K. pneumoniae to povidone-iodine, an increase in the sensitivity of bacteria to antibiotics was shown. The minimum inhibitory concentrations (MIC) of 11 antibacterial agents in relation to the studied cultures decreased by an average of 2.35–23.2 times compared with the control values. Under the experimental conditions, the sensitivity of the studied cultures increased to amoxicillin/clavulanic acid by ≥ 2 – ≥ 8 times, piperacillin/tazobactam — by ≥ 2 – ≥ 32 times, ceftazidime — by 4 – ≥ 16 times, ami-kacin — 2 – ≥ 16 times, ertapenem — 2.0–8.0 times, meropinem — 2–8 times, cefepim — ≥ 4 – ≥ 64 times, ciprofloxacin — 4 – ≥ 16 times, gentamicin — 2 – ≥ 8 times, norfloxacin — 2–8 times, nitrofurantoin — 2–4 times. An increase in the level of sensitivity to antibiotics was found in 50–100% of the studied clinical bacterial isolates. A similar effect has not been established for ampicillin, cefotaxime, imipenem, and trimethoprim/sulfamethoxazole. It should be noted that under the conditions of the experiment, intraspecific heterogeneity of clinical isolates of K. pneumoniae was revealed in terms of the level of acquired sensitivity to antibacterial agents.
Discussion. Our results are not described in the scientific literature and require further study and explanation.
 209-218
209-218


REVIEWS
The Russian database of HIV antiretroviral drug resistance
Abstract
The development of sequencing technologies and bioinformatic analysis made it possible to conduct molecular and epidemiological studies, in which nucleotide sequences of the human immunodeficiency virus (HIV) are used as information added to the patient profile. From a practical perspective, studies of prevalence of HIV drug resistance (HIVDR) are of the highest significance. To promote such studies, different countries use databases that serve as repositories of genetic and epidemiological information. The Russian HIVDR database (https://hivresist.ru/) was created in 2009. Nevertheless, it was characterized by limited applicability for a long time. Since 2021, after the regulatory documents had been revised and updated, the entry of HIVDR research results into the Russian HIVDR database has been mandatory. Therefore, the priority attention has been given to upgrading the database and improving its functional capabilities. Different methods have been developed to enter clinical, epidemiological and genetic data. At the time of this study, the Russian database HIVDR contained 10,626 unique records about patients and 13,126 nucleotide sequences deposited by 10 institutions. The following functions have been provided for data analysis: quality control of the epidemiological and clinical information about a patient, quality control of nucleotide sequences, contamination check, subtyping, detection of DR mutations, identification of viral tropism and generation of standardized reports. The efforts toward further development of the Russian HIVDR database will be focused on designing tools for detection and analysis of molecular clusters, adaptation to routine application for epidemiological surveillance of HIV infection.
 219-227
219-227


The Yersinia pestis aggregation as a functional adaptation to the flea organism (review)
Abstract
The understanding of mechanisms and conditions of plague microbe aggregation in vectors’ (fleas) organisms has been of interest for a long time, and, with start of biofilm research in 1990s, it got a new perspectives.
With the aim to determine the characteristics of the plague microbe aggregation in the organism the main vector in Tuva natural plague focus’ — flea of Citellophilus tesquorum species, the data of many years of experimental studies focused on typical virulent strains of Yersinia pestis subsp. pestis were summarized and analyzed. Ectoparasites were infected and fed on long-tailed ground squirrel (Spermophilus undulatus), which is their natural host and main carrier of plague in Tuva natural plague focus. Interaction between Y. pestis and fleas were estimated using the rate of individuals with conglomerates “bacterial lumps”, formed during feeding, and “blocks” in the proventriculus foRmed during feeding and throughout the whole experiment registered in alive fleas after bloodsucking, and also using frequency of causative agent transmission to animals used for feeding.
The analysis identified factors influencing the frequency and dynamics of different Y. pestis aggregate types forming into C. tesquorum, and suggest that these processes are functional adaptation of microbe to the flea organism.
 240-250
240-250


Immunogenesis in Lassa fever and prospects for vaccine development
Abstract
Analysis of literature data on the structure of the Lassa virus genome and its strain diversity has shown that the molecular heterogeneity of strains is essential in the design of vaccines and evaluation of their efficacy, which is determined by the relevant WHO recommendations. During virus reproduction, Lassa counteracts cellular immunogenesis by suppressing the expression of the suppressor of signaling proteins, cytokines and the RLR receptor that recognizes viral two-segmented RNA.
The GP protein, which determines the pathogen's infectivity and tropism, should be the main target for vaccines being developed. Other targets are the processes of viral RNA synthesis that determine the features of immunogenesis. A study of the immunogenesis of Lassa fever shows that the preferred candidate would be a replicating apathogenic vaccine capable of inducing an optimal combination of cellular and humoral responses, that is, causing high activity of T-lymphocytes and the production of viral neutralizing antibodies. One of the most important characteristics of a live candidate vaccine should be genetic stability to exclude reversion to a more pathogenic genotype.
Of the more than 130 candidate vaccines against Lassa fever designed, only two being the most promising were tested for immunogenicity and safety in humans (a recombinant on the measles virus platform and a DNA vaccine). A recombinant of Lassa virus with vesicular stomatitis virus and reassortants of Mopeya and Lassa viruses (MOPV/LASV- ML29) and (r3ML29) are promising for further development. A candidate vaccine rVSVΔG/LVGPC, similar in design to the rVSVΔG-ZEBOV-GP vaccine against Ebola, is promising.
 228-239
228-239


OBITUARIES
 251-252
251-252