The structure of ESKAPE pathogens isolated from patients of the neonatal intensive care unit at the National Hospital of Pediatrics in Hanoi, the Socialist Republic of Vietnam
- Authors: Stepanova T.F.1, Kataeva L.V.1, Posoyuznykh O.V.1, Bogun A.G.2, Kislichkina A.A.2, Tran T.N.3
-
Affiliations:
- Tyumen Region Infection Pathology Research Institute
- State Scientific Center of Applied Microbiology and Biotechnology
- Joint Russian-Vietnamese Tropical Research and Technology Center (Tropical Center)
- Issue: Vol 100, No 2 (2023)
- Pages: 168-177
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/11141
- DOI: https://doi.org/10.36233/0372-9311-329
- EDN: https://elibrary.ru/aamnei
- ID: 11141
Cite item
Abstract
Introduction. The incidence of healthcare-associated infections is a major public health problem worldwide, affecting all countries regardless of their economic status. The main agents of these infections are pathogens belonging to the ESKAPE group.
The aim of the study was to explore the structure, molecular and antigenic characteristics of the ESKAPE pathogens isolated from oral and anal mucosa of patients of the neonatal intensive care unit (NICU), and to assess their etiological significance in occurrence of healthcare-associated infections.
Materials and methods. Samples from a total of 49 children were tested, including 40 newborns – patients of NICU at the National Hospital of Pediatrics in Hanoi. Collection and processing of biomaterial (oropharyngeal swabs, rectal swabs) and isolation of bacterial cultures were performed using conventional bacteriological methods. Mass spectrometry was used for identification of isolates. Klebsiella pneumoniae strains were analyzed using the whole-genome sequencing method.
Results. The group of gram-positive ESKAPE pathogens identified in oral mucosa was represented by isolates Enterococcus faecium and Staphylococcus aureus. The isolates of the family Enterobacteriaceae included K. pneumoniae, Escherichia coli, Enterobacter cloacae; the group of nonfermenting gram-negative bacteria was represented by Pseudomonas aeruginosa, Acinetobacter baumannii. The structure of ESKAPE pathogens persistent in anal mucosa was characterized by dominance of Enterococcus spp., E. coli, K. pneumoniae and P. aeruginosa bacteria. The whole-genome sequencing of K. pneumoniae isolates revealed 7 clusters and 8 sequence types. ST14 and ST1741 prevailed, accounting for 25%, respectively, of the total number of the studied strains. The molecular serotyping showed that by the O antigen, strains belonged mainly to serotypes O1v1, O1/ O2v2, O5; by the presence of the capsular antigen — to serotypes KL2, KL104, KL60.
Conclusion. The analysis of the structure of the ESKAPE pathogens isolated from the oral and anal mucosa of patients of NICU at the National Hospital of Pediatrics in Hanoi identified etiologically significant agents of bacterial infections: S. aureus, K. pneumoniae, E. coli, E. cloacae, P. aeruginosa, A. baumannii. The molecular and genetic analysis of K. pneumoniae strains co-circulating in mucous membranes of several patients of the unit revealed their homology, thus confirming healthcare-associated contamination of children with nosocomial strains.
Full Text
Introduction
Neonatal bacterial infections are associated with multiple risk factors. The diversity of clinical forms of childhood infections is explained, first of all, by immaturity of natural barrier defenses, especially, when antimicrobial therapy is involved [1]. The level of infectious morbidity is determined mainly by the frequency of damage to the skin, eyes, navel and gastrointestinal tract. Therefore, monitoring of circulating ESKAPE pathogens is of critical importance at maternity care centers, as the risk of bacterial diseases is increased due to the physiological status of newborns [2, 3].
Widespread occurrence of healthcare-associated infections (HAIs) in any healthcare facility, most particularly, in maternity and pediatric hospitals causes significant damage to the health of newborns and general population, economy and demographic situation in various countries [4, 5]. Frequently, the numbers of cases of neonatal hospital-acquired infections, which are reported in Russian and foreign publications, cannot be compared due to the absence of common approaches to the system of recording and reporting diseases [6, 7].
The spread of multidrug-resistant nosocomial pathogens causing infectious complications in children raises concerns among clinicians worldwide. In recent years, the ESKAPE group has been highlighted among the etiologically significant bacteria, including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp. The dominant role among these agents of hospital-acquired infections belongs to the pathogens that are characterized by diversity of antimicrobial resistance mechanisms. The World Health Organization included ESKAPE pathogens in the group of microorganisms of the highest priority for development of new antibiotics or new treatment methods [8].
The burden of hospital-acquired infections, especially among children, in low and middle-income countries is unknown, mostly due to a lack of reported data. The European multicenter study of cases with hospital-acquired infections in pediatric hospitals showed that their prevalence ranged from 2.5% in general care units to 23.6% in intensive care units. High rates of these infections were also recorded in neonatal intensive care units, where their prevalence was 3–20 times as high in resource-limited settings compared to high-income countries.
Vietnam is a low-income country with fast growing public and private health sectors. Because of the low levels of the gross national income and national health expenditure per capita, personal health care expenditures account for around 80%. Antibiotics are widely used by the population and are most commonly purchased in private pharmacies and clinics. The healthcare sector is overloaded with the relatively low number of healthcare personnel per capita (7 doctors per 10,000 population), shortage of inpatient beds and hospital overcrowding reaching 170%. Vietnam has no national system of epidemiological surveillance of HAIs; the data on cases in intensive care units are limited. Considering high infection rates, Vietnam becomes a "hotspot" for the emergence of antimicrobial resistance with the rates among the highest in Asia [9–11].
The earlier performed studies of hospital-acquired infections in intensive care units at 3 Vietnamese pediatric hospitals showed that their prevalence was 33% [9]. K. pneumoniae is among the leading bacteria that cause nosocomial infections and can develop antimicrobial resistance [11–13]. The results of molecular and genetic studies of strains of bacteria co-circulating in a particular unit of a specific hospital show their high heterogeneity. Priority attention should be given to measures preventing the emergence of highly virulent and resistant strains and stopping their spread [14].
The study of the diversity of ESKAPE pathogens contaminating newborns, including premature infants receiving treatment in neonatal intensive care units (NICU) is a top-priority task of healthcare organizations both in Russia and other countries; its achievement will help minimize the risks associated with adverse epidemiological situations.
The aim of the study was to explore the structure, molecular and antigenic characteristics of ESKAPE pathogens isolated from oral and anal mucous membranes of NICU patients, and to assess their etiological significance in occurrence of HAIs.
The studies were conducted in accordance with R&D of the Tyumen Research Institute for Regional Infectious Diseases Pathology of Rospotrebnadzor in the pursuance of the Action Plan for Provision of Scientific, Methodological, Material and Technical Assistance to the Socialist Republic of Vietnam, addressing the threat of infectious diseases and minimization of risks associated with health-hazardous chemical substances in 2020–2022.1
Materials and methods
The study included 49 children aged 0–60 days, including 40 newborns (the median was 13 days; Q1–Q3: 7–23), who received treatment in NICU at the National Hospital of Pediatrics in Hanoi. Biomaterial samples were collected simultaneously from all children, regardless of their age, type of delivery, degree of prematurity, birth weight, type of feeding, length of stay in the unit, antibiotic treatment (the random sampling method). The study was performed with the voluntary informed consent of representatives of the patients. The study protocol was approved by Ethics Committee of the Tyumen Research Institute for Regional Infectious Diseases Pathology of Rospotrebnadzor (minutes No. 1 of 01.06.2022).
The bacteriological study was performed using 98 samples of biomaterial collected from oral and anal mucous membranes of the children; microbial growth was recorded in 96 specimens. A total of 101 isolates were isolated from the oral mucosa of the children, and 137 isolates were isolated from the anal mucosa. The K. pneumoniae strains (n = 20) isolated from the above loci were studied using whole-genome sequencing.
Microbiological studies were conducted using the conventional bacteriological method. The species identification of bacteria was confirmed by matrix-assisted laser desorption ionization mass spectrometry using the Maldi BioTyper 3.0 software. The identification level above 2.0 was indicative of a high degree of confidence.
The obtained reads for each strain were assembled into contigs using the Unicycler v.0.4.7 program. Samples with the average genome coverage above 200 indicated that the amount of data was sufficient. The results of whole-genome sequencing were analyzed using the Kraken Metagenomics version 2 software for analysis of metagenomic samples (the classifier of reads — short nucleotide sequences). Phylogenetic studies were performed using the Wombac 2.0 program that made it possible to find core single nucleotide polymorphisms (SNPs) in nucleotide sequences and to perform alignment of these polymorphisms. The Kaptive web-based tool was used for detection of O antigens and capsular antigens of K. pneumoniae strains. Sequence types were identified using MLST 2.02 and Lasergene software.
Microsoft Office Excel 2016 spreadsheets were used for collecting, correcting, systematizing of the input information and for visualization of the obtained results. Statistical methods and the SPSS v. 22 software were used for analysis of the received data. Nominal data were described using absolute values and percentages, including 95% confidence intervals (CI) — the Clopper–Pearson method.
Results
The results of the bacteriological studies of the children’s oral and anal mucosa show that the obtained isolates are represented by gram-positive and gram-negative bacteria as well as by the minor quantity of fungi of the genus Candida. The group of gram-positive bacteria (27.3% [95% CI, 21.7–33.4]) was comprised by bacteria of the genera Staphylococcus (S. aureus and other species), Streptococcus (primarily S. vestibularis, S. salivarius) and Enterococcus faecalis. Gram-negative isolates are represented by Enterobacterales bacteria (41.6% [95% CI, 35.3–48.1]) and nonfermenting gram-negative bacteria (22.7% [95% CI, 17.5–28.5]). Among representatives of Enterobacterales, we identified bacteria of the following genera: Escherichia, Klebsiella, Serratia, Enterobacter; the cluster of nonfermenting gram-negative bacteria was comprised by Acinetobacter spp., Pseudomonas spp., Stenotrophomonas maltophilia and Elizabethkingia meningoseptica. The isolates from the studied loci demonstrated the dominance of Enterobacterales bacteria. Fungi of the genus Candida accounted for 8.4% [95% CI, 5.2–12.7].
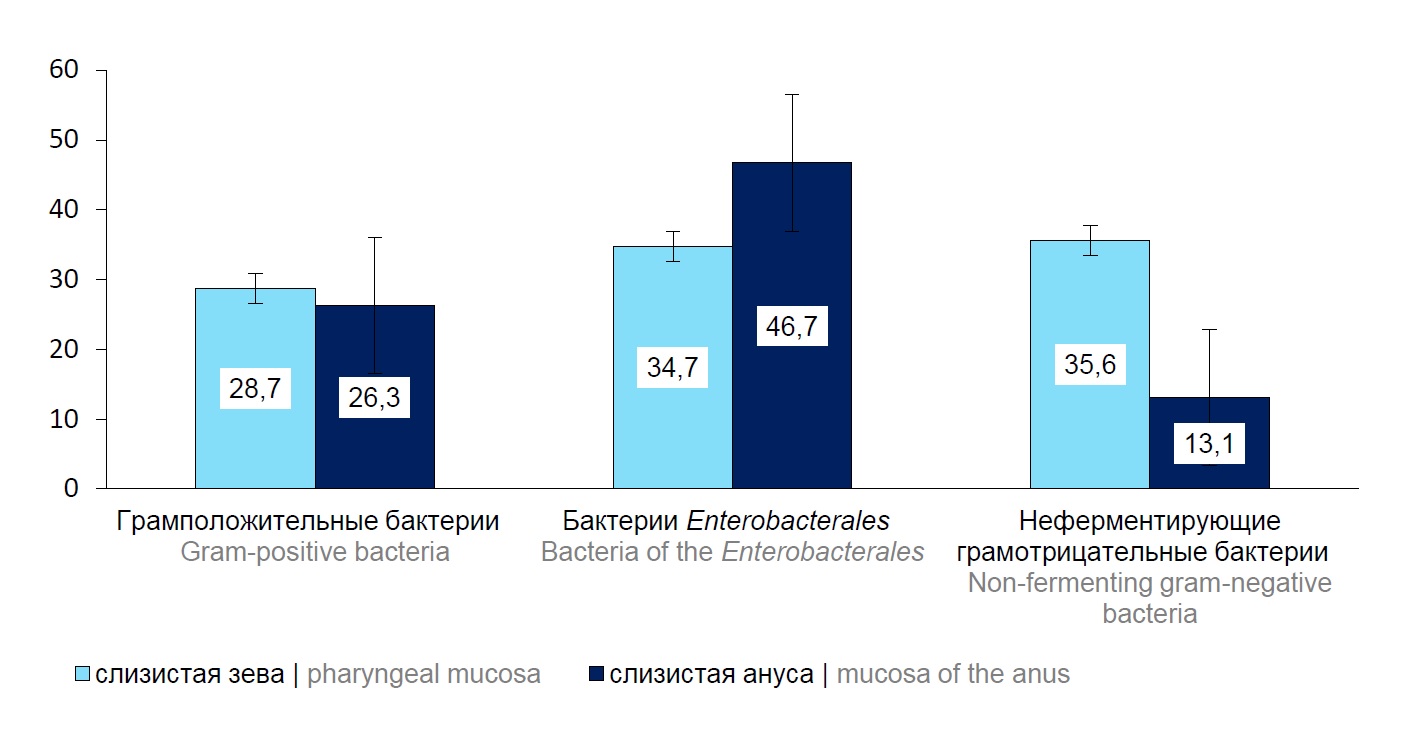
The analysis of the quantity of the isolates from the specified loci showed insignificant dominance of Enterobacterales bacteria in anal mucosa. In the children’s oral mucosa, nonfermenting gram-negative bacteria were detected in 35.6% of cases [95% CI, 26.4–45.8] compared to their detection rate for anal mucosa — 13.1% [95% CI, 8.0–19.9] (Fig. 1). Groups of gram-positive bacteria isolated from oral and anal mucosa were detected almost in equal percentages.
Fig. 1. The detection frequency of isolates of the main bacteria groups in oral and anal mucosa of the children, %.
Among the total number of identified isolates from the specified loci, 51.7% [95% CI, 45.1–58.2] of strains belonged to the ESKAPE group of pathogens: S. aureus, K. pneumoniae, E. coli, E. cloacae, P. aeruginosa, A. baumannii. The comparative analysis of the structure of ESKAPE bacteria identified from the biomaterial samples collected from oral and anal mucosa of the children is presented in Fig. 2.
Fig. 2. Comparative analysis of ESKAPE pathogens isolated from oral and anal mucosa of the children, %.
The analysis of isolates of the ESKAPE group, which were obtained from the oral mucosa, showed that bacteria of the family Enterobacteriaceae and nonfermenting gram-negative bacteria prevailed in the above locus. At the same time, S. viridans and coagulase-negative Staphylococcus spp., which represent a resident microbiota of the oral cavity, were identified in oral mucous membranes of the examined children.
The bacteriological analysis of the samples of biological materials collected from the patients showed that K. pneumoniae bacteria prevailed in microbiota of the oral and rectal mucosa in 51.9% of cases. The results of whole-genome sequencing of K. pneumoniae strains isolated from the oral mucosa (n = 9) and the rectal mucosa (n = 11) show that the selected strains were divided into 7 genetic clusters. Cluster I included K. pneumoniae strains 39, 51, 57, 85 (isolated from the oral mucosa) and strains 42, 56, 96 (from the rectal mucosa). The structural analysis of core SNP genomes of K. pneumoniae strains within cluster I showed that the cluster is divided into two groups with genomes differing from each other by a little over 10,000 SNP. The first group is represented by strains 39, 42, 51, 56, 57; the second group — by strains 85, 96. The results of pairwise comparison of contig assemblies are shown in Table 1. The analysis of core SNPs in the first group of strains showed that they differed not more than by 55, thus confirming the high homology of these isolates.
Table 1. Results of comparison of contig assemblies of K. pneumoniae strains belonging to cluster I
SNP-dists 0.6.3 | Strain 85 | Strain 96 | Strain 51 | Strain 56 | Strain 57 | Strain 39 |
K. pneumoniae 96 | 19 | |||||
K. pneumoniae 51 | 10987 | 10987 | ||||
K. pneumoniae 56 | 10997 | 10993 | 52 | |||
K. pneumoniae 57 | 10980 | 10980 | 17 | 55 | ||
K. pneumoniae 39 | 10986 | 10984 | 15 | 55 | 16 | |
K. pneumoniae 42 | 10986 | 10986 | 11 | 51 | 8 | 10 |
Cluster II was formed by strains 91, 49 (from the oral mucosa) and strains 92, 50, 66 (from the rectal mucosa); cluster III — by strains 53 (from the oral mucosa) and strains 54, 68 (from the rectal mucosa). Clusters IV, V, VI, and VII are represented by strains 74 and 82 — from the rectal mucosa and strains 59, 31 — from the oral mucosa. The molecular serotyping by O and capsular antigens showed that each cluster of K. pneumoniae strains could be assigned to a specific serotype (Table 2). By the O antigen, all strains of clusters I and VII are assigned to serotype Olvl. Strains of the second cluster had some differences by the O antigen and were assigned to Ol/O2v2 and Olv2, including the strain of cluster V. Clusters III and IV were represented by strains containing the O5 antigen. The strain with the O3b antigen was assigned to cluster VI.
Table 2. Results of molecular serotyping of K. pneumoniae strains
Name of the strain | KL-antigen | О-antigen | Cluster |
K. pneumoniae 39 | KL2 | Olvl | I |
K. pneumoniae 42 | KL2 | Olvl | |
K. pneumoniae 51 | KL2 | Olvl | |
K. pneumoniae 56 | KL2 | Olvl | |
K. pneumoniae 57 | KL2 | Olvl | |
K. pneumoniae 85 | KL112 | Olvl | |
K. pneumoniae 96 | KL112 | Olvl | |
K. pneumoniae 92 | KL104 | Ol/O2v2 | II |
K. pneumoniae 50 | KL104 | Ol/O2v2 | |
K. pneumoniae 66 | KL104 | Olv2 | |
K. pneumoniae 49 | KL104 | Ol/O2v2 | |
K. pneumoniae 91 | KL104 | Ol/O2v2 | |
K. pneumoniae 68 | KL60 | O5 | III |
K. pneumoniae 54 | KL60 | O5 | |
K. pneumoniae 53 | KL60 | O5 | |
K. pneumoniae 74 | KL114 | O5 | IV |
K. pneumoniae 59 | KL62 | Olv2 | V |
K. pneumoniae 82 | KL128 | O3b | VI |
K. pneumoniae 31 | KL62 | Olvl | VII |
K. pneumoniae 95 | – | – | – |
Note. Strain 95 — non-serotyped by the O antigen and KL antigen.
The analysis of the capsular antigen (KL) showed that strains of all clusters were homogenous, except for cluster I, in which 5 K. pneumoniae strains belonged to serotype KL2 and 2 strains belonged to KL112.
The multilocus sequence typing revealed that strains of a specific cluster belonged to the same sequence type (ST), except for cluster I that included ST14 and ST15. Regarding the homology of strains by clusters, it was found that genotypes in all clusters were identical by loci tonB, rpoB, phoE, pgi, mdh, gapA; differences were recorded only in cluster I, locus infB (Table 3).
Table 3. Results of multilocus sequence typing of K. pneumoniae strains
Name of the strain | Sequence type | Loci | Cluster | ||||||
tonB | rpoB | phoE | pgi | mdh | gapA | infB | |||
K. pneumoniae 39 | 14 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | I |
K. pneumoniae 42 | 14 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |
K. pneumoniae 51 | 14 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |
K. pneumoniae 56 | 14 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |
K. pneumoniae 57 | 14 | 1 | 1 | 1 | 1 | 1 | 1 | 6 | |
K. pneumoniae 85 | 15 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
K. pneumoniae 96 | 15 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
K. pneumoniae 92 | 1741 | 42 | 4 | 17 | 1 | 4 | 2 | 3 | II |
K. pneumoniae 50 | 1741 | 42 | 4 | 17 | 1 | 4 | 2 | 3 | |
K. pneumoniae 66 | 1741 | 42 | 4 | 17 | 1 | 4 | 2 | 3 | |
K. pneumoniae 49 | 1741 | 42 | 4 | 17 | 1 | 4 | 2 | 3 | |
K. pneumoniae 91 | 1741 | 42 | 4 | 17 | 1 | 4 | 2 | 3 | |
K. pneumoniae 68 | 1224 | 169 | 38 | 142 | 63 | 26 | 18 | 67 | III |
K. pneumoniae 54 | 1224 | 169 | 38 | 142 | 63 | 26 | 18 | 67 | |
K. pneumoniae 53 | 1224 | 169 | 38 | 142 | 63 | 26 | 18 | 67 | |
K. pneumoniae 74 | 3918, 1928, 4038 | 124 | 17 | 41 | 33 | 21 | 28 | 18 | IV |
K. pneumoniae 59 | 730 | 56 | 4 | 1 | 1 | 33 | 2 | 1 | V |
K. pneumoniae 82 | 16 | 4 | 4 | 4 | 1 | 2 | 2 | 1 | VI |
K. pneumoniae 31 | 48 | 10 | 1 | 7 | 2 | 2 | 2 | 5 | VII |
K. pneumoniae 95 | – | – | – | – | – | – | – | – | – |
Discussion
The examined children were patients in the intensive care unit of a large specialized pediatric center and most of them were premature infants. The degree of prematurity of newborns depended on perinatal risk factors: type of delivery, gestational age, birth weight, asphyxia, respiratory distress syndrome or congenital abnormalities. Although the above data were not provided for the analysis, they can contribute to predisposition to contamination of children’s mucous membranes with ESKAPE pathogens circulating in the unit and to development of bacterial diseases.
The studies of oral and rectal mucous membranes of NICU patients at the National Hospital of Pediatrics in Hanoi demonstrated a great diversity of circulating bacteria belonging to the ESKAPE group and being etiologically significant agents of bacterial infections, including HAIs. Note that K. pneumoniae and P. aeruginosa isolates from rectal mucosa belong to highly dangerous pathogens and are responsible for most of the hospital-acquired infections in immunocompromised patients [15, 16]. At the same time, Enterococcus spp. and E. coli bacteria are important components of gut microbiota [13, 17].
Some publications addressing cases of HAIs in intensive care units at Vietnamese pediatric hospitals state that K. pneumoniae bacteria are the most frequent pathogen isolated from patients. Nosocomial strains of these bacteria characterized by a wide range of virulence and resistance genes prevail in hospitals in Europe, the United States, East and Southeast Asia [18–21].
Molecular and genetic studies of the microbiome help evaluate the pathogenic potential of microbial communities living in certain niches. The polysaccharide chain composing the structure of the outer membrane of K. pneumoniae possesses antigenic properties and helps divide strains into O serotypes. By the O antigen, all strains of 7 clusters had some differences and were assigned to serotypes O1v1, Ol/O2v2 and Olv2, O3b and O5. At the same time, 70% of the strains had antigens of serotype O1. There are data confirming that serotypes O1, O2, O3 are the most prevalent and are detected in 80% of cases of infections caused by K. pneumoniae bacteria [22]. The analysis of the capsular antigen showed that strains of all clusters were also characterized by diversity with detected serotypes KL2, KL60, KL62, KL104, KL112, KL114 and KL128. Serotypes KL2 and KL104 were detected in half of the strains (25% respectively). In clinical settings, hypervirulent serotypes KL1 and KL2 should be given close attention. They are characterized by a hypermucoid phenotype and are associated with especially severe Klebsiella infection [23].
The multilocus sequence typing demonstrated the homology of strains by clusters; genotypes in all clusters were identical by loci tonB, rpoB, phoE, pgi, mdh, gapA; differences were recorded in locus infB. It was found that sequence types ST14, ST1741 and ST1224 prevailed. In addition, two strains were assigned to sequence type ST15. There is evidence suggesting prevalence of ST11 strains in East Asia; ST15 strains occur more rarely, though worldwide they are reported as the source of nosocomial outbreaks of resistant K. pneumoniae [12, 20]. Research publications report that epidemiological conditions in pediatric hospitals are associated with the circulation of K. pneumoniae strains belonging to sequence types ST17, ST3181 and ST1564 [24].
Thus, the performed molecular and genetic studies demonstrate that the nosocomial population of K. pneumoniae isolates was characterized by the co-circulation of homologous strains in mucous membranes of several patients in the unit, thus confirming that the newborns were contaminated with them while receiving treatment. The analysis of species composition of circulating microorganisms, whole-genome sequencing, serotyping and multilocus sequence typing as well as identification of genetic markers of the pathogenic potential of strains are essential for effective identification of HAI pathogens [25].
The etiological structure of infections, including HAIs, depends on the conditions in a specific unit of a healthcare facility. Therefore, monitoring of the circulation of pathogens belonging to the ESKAPE group is required not only when the occurrence of bacterial infection has been reported, but also routinely [26]. The microbiological monitoring of ESKAPE pathogens, which is based on molecular and genetic studies, will be instrumental in detection of HAI pathogens and forecasting of epidemiological situations when providing care to newborns.
Ethics approval. The study was performed with the voluntary informed consent of the legal representatives of underage patients. The research protocol was approved by the Ethics Committee of the Tyumen Region Infection Pathology Research Institute (protocol No. 1, June 1, 2022).
1 Executive Order No. 1536-p of the Government of the Russian Federation, 13/7/2019.
2 Multi-Locus Sequence Typing.
URL: https://cge.cbs.dtu.dk/services/MLST
About the authors
Tatyana F. Stepanova
Tyumen Region Infection Pathology Research Institute
Email: info@tniikip.rospotrebnadzor.ru
ORCID iD: 0000-0002-6289-6274
D. Sci. (Med.), Professor, Director
Russian Federation, TyumenLyubov V. Kataeva
Tyumen Region Infection Pathology Research Institute
Author for correspondence.
Email: info@tniikip.rospotrebnadzor.ru
ORCID iD: 0000-0001-9966-8454
D. Sci. (Med.), chief researcher, Head, Bacteriological laboratory
Russian Federation, TyumenOlga V. Posoyuznykh
Tyumen Region Infection Pathology Research Institute
Email: info@tniikip.rospotrebnadzor.ru
ORCID iD: 0000-0002-4364-9868
biologist, Bacteriological laboratory
Russian Federation, TyumenAlexander G. Bogun
State Scientific Center of Applied Microbiology and Biotechnology
Email: info@tniikip.rospotrebnadzor.ru
ORCID iD: 0000-0001-5454-2495
Cand. Sci. (Biol.), Head, Department of collection cultures
Russian Federation, ObolenskAngelina A. Kislichkina
State Scientific Center of Applied Microbiology and Biotechnology
Email: info@tniikip.rospotrebnadzor.ru
ORCID iD: 0000-0001-8389-2494
Cand. Sci. (Biol.), senior researcher, Department of collection cultures
Russian Federation, ObolenskThi Nhai Tran
Joint Russian-Vietnamese Tropical Research and Technology Center (Tropical Center)
Email: info@tniikip.rospotrebnadzor.ru
ORCID iD: 0009-0003-1892-3241
researcher
Viet Nam, HanoiReferences
- Mukhamedova Sh.T., Shamsutdinov A.S., Khamraeva D.R., Karomatova F.A. Nosocomial infections in newborns. Biologiya i integrativnaya meditsina. 2021;(3):75–86. EDN: https://www.elibrary.ru/eosqum
- Nikitina I.V., Gerasimova A.V., Ivanova L.A., et al. Health care-associated infections in critically ill premature newborns: epidemiology, clinical features and diagnostics in modern conditions. Neonatology: News, Opinions, Training. 2020;8(3):7–17. DOI: https://doi.org/10.33029/2308-2402-2020-8-3-7-17 EDN: https://www.elibrary.ru/ikcmfj
- Martin R.M., Bachman M.A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018;8:4. DOI: https://doi.org/10.3389/fcimb.2018.00004
- Bogdanova N.M. Prevention of intrabolichic infection in general supporting institutions at the organization of breastfeeding. Medicine: Theory and Practice. 2019;4(1):132–7. EDN: https://www.elibrary.ru/klykty
- Ivanova M.V., Mindlina A.Ya., Polibin R.V., Ushanova A.V. Russia-wide epidemiological survey of congenital and nosocomial infections in newborns. Russian Journal of Infection and Immunity. 2019;9(1):193–202. DOI: https://doi.org/10.15789/2220-7619-2019-1-193-202 EDN: https://www.elibrary.ru/zgxqvf
- Naigovzina N.B., Popova A.Yu., Biryukova E.E., et al. Optimization of the system of measures to control and prevent infections associated with the provision of medical care in the Russian Federation. Healthcare Management: News, Views, Education. Bulletin of VSHOUZ. 2018;(1):17–26. EDN: https://www.elibrary.ru/yusipb
- Naumkina E.V., Pakhalkova E.V., Pyadochkina T.V., Abrosimova O.A. Microbiological monitoring in the departments of the perinatal center. Mother and Baby in Kuzbass. 2018;(1):23–7. EDN: https://www.elibrary.ru/yopgqq
- Yarets Yu.I. Pathogenic potential of ESKAPE group bacteria isolated from wounds: characterization of phenotypic and genotypic markers and possibility of their practical application. Journal of the Grodno State Medical University. 2022;20(4):400–13. DOI: https://doi.org/10.25298/2221-8785-2022-20-4-400-413 EDN: https://www.elibrary.ru/vtsale
- Le N.K., Hf W., Vu P.D., et al. High prevalence of hospital-acquired infections caused by gram-negative carbapenem resistant strains in Vietnamese pediatric ICUs: A multi-centre point prevalence survey. Medicine (Baltimore). 2016;95(27):e4099. DOI: https://doi.org/10.1097/md.0000000000004099
- Berglund B., Hoang N.T.B., Lundberg L., et al. Clonal spread of carbapenem-resistant Klebsiella pneumoniae among patients at admission and discharge at a Vietnamese neonatal intensive care unit. Antimicrob. Resist. Infect. Control. 2021;10(1):162. DOI: https://doi.org/10.1186/s13756-021-01033-3
- Berglund B., Hoang N.T.B., Tärnberg M., et al. Colistin- and carbapenem-resistant Klebsiella pneumoniae carrying mcr-1 and blaOXA-48 isolated at a paediatric hospital in Vietnam. J. Antimicrob. Chemother. 2018;73(4):1100–2. DOI: https://doi.org/10.1093/jac/dkx491
- Piperaki E.T., Syrogiannopoulos G.A., Tzouvelekis L.S., Daikos G.L. Klebsiella pneumoniae: virulence, biofilm and antimicrobial resistance. Pediatr. Infect. Disease. 2017;36(10):1002–5. DOI: https://doi.org/10.1097/inf.0000000000001675
- Bondarenko A.P., Trotsenko O.E., Zaitseva T.A., et al. Microbiological and molecular-biological methods in epidemiological evaluation of healthcare associated infections caused by Klebsiella pneumoniae. The Far Eastern Journal of Infectious Pathology. 2021;(41):68–75. EDN: https://www.elibrary.ru/ypthje
- Skachkova T.S., Shipulina O.Yu., Shipulin G.A., et al. Characterization of genetic diversity of the Klebsiella pneumoniae strains in a Moscow tertiary care center using next-generation sequencing. Clinical Microbiology and Antimicrobial Chemotherapy. 2019;21(1):69–74. EDN: https://www.elibrary.ru/wuxmap
- Zheng X., Wang J.F., Xu W.L., et al. Clinical and molecular characteristics, risk factors and outcomes of Carbapenem-resistant Klebsiella pneumoniae bloodstream infections in the intensive care unit. Antimicrob. Resist. Infect. Control. 2017;6:102. DOI: https://doi.org/10.1186/s13756-017-0256-2
- Belova I.V., Tochilina A.G., Solov'eva I.V., et al. Characteristic of hospital Klebsiella pneumoniae strains circulating in the pediatric hospital. Public Health and Life Environment – PH&LE. 2019;(8):25–9. DOI: https://doi.org/10.35627/2219-5238/2019-317-8-25-29 EDN: https://www.elibrary.ru/bbavei
- Peters L., Olson L., Khu D.T.K., et al. Multiple antibiotic resistance as a risk factor for mortality and prolonged hospital stay: A cohort study among neonatal intensive care patients with hospital-acquired infections caused by gram-negative bacteria in Vietnam. PLoS One. 2019;14(5):e0215666. DOI: https://doi.org/10.1371/journal.pone.0215666
- Berglund B., Hoang N.T.B., Tärnberg M., et al. Molecular and phenotypic characterization of clinical isolates belonging to a KPC-2-producing strain of ST15 Klebsiella pneumoniae from a Vietnamese pediatric hospital. Antimicrob. Resist. Infect. Control. 2019;16(8):156. DOI: https://doi.org/10.1186/s13756-019-0613-4
- Fursova N.K., Astashkin E.I., Lev A.I., et al. Phenotypes and genotypes of classical and hypervirulent Klebsiella pneumoniae clinical strains isolated in Moscow in 2013–2018. Russian Journal of Infection and Immunity. 2018;8(4):747. EDN: https://www.elibrary.ru/pohodv
- Chebotar' I.V., Bocharova Yu.A., Podoprigora I.V., Shagin D.A. The reasons why Klebsiella pneumoniae becomes a leading opportunistic pathogen. Clinical Microbiology and Antimicrobial Chemotherapy. 2020;22(1):4–19. DOI: https://doi.org/10.36488/cmac.2020.1.4-19 EDN: https://www.elibrary.ru/ockpac
- Yu V.L., Hansen D.S., Ko W.C., et al. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg. Infect. Dis. 2007;13(7):986–93. DOI: https://doi.org/10.3201/eid1307.070187
- Tochilina A.G., Belova I.V., Solov'eva I.V., et al. Molecular typing of hospital strains of Klebsiella pneumoniae ssp. pneumoniae. Clinical Microbiology and Antimicrobial Chemotherapy. 2019;21(S1):63–4. EDN: https://www.elibrary.ru/ikfgvt
- Kuz'menko S.A., Brezhneva N.I., Goncharov A.E., Tutel'yan A.V. Features of nosocomial Klebsiella pneumoniae population. Fundamental and Clinical Medicine. 2019;4(2): 58–65. DOI: https://doi.org/10.23946/2500-0764-2019-4-2-58-65 EDN: https://www.elibrary.ru/fwfxlj
- Kondratenko T.A., Sheozheva A.V. Monitoring of the microbiocenosis in newborns within intensive care units. Preventive and Clinical Medicine. 2018;(2):31–3. EDN: https://www.elibrary.ru/xwtohb
Supplementary files