Molecular and biological characterization of Streptococcus pneumoniae isolates from patients with pneumococcal meningitis
- Authors: Chagaryan A.N.1, Ivanchik N.V.1, Kuzmenkov A.Y.1, Kozlov R.S.1,2, Gaponova I.I.2, Mironov K.O.2
-
Affiliations:
- Research Institute of Antimicrobial Chemotherapy, Smolensk State Medical University
- Central Research Institute of Epidemiology
- Issue: Vol 102, No 2 (2025)
- Pages: 150-161
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18823
- DOI: https://doi.org/10.36233/0372-9311-614
- EDN: https://elibrary.ru/NRAEKS
- ID: 18823
Cite item
Abstract
The aim of the study is to provide key characteristics of Streptococcus pneumoniae isolates circulating in Russia in 2015–2020 and isolated from pneumococcal meningitis patients based on high-throughput sequencing data, including global pneumococcal sequence clusters, serotypes, virulence factors and genetic determinants of resistance, in comparison with clinical data on antimicrobial susceptibility.
Materials and methods. We studied 68 invasive S. pneumoniae isolates from blood and cerebrospinal fluid of patients with bacterial meningitis in different regions of Russia in 2015–2020. Species identification was performed taking into account the morphology of colonies on blood agar, the presence of α-hemolysis, negative catalase reaction, sensitivity to optoquine, and positive latex-agglutination results. The sensitivity of isolates to antimicrobials was determined by microdilution in broth, and sensitivity categories were determined based on borderline values of minimum inhibitory concentrations (MICs). Whole genome sequencing of S. pneumoniae isolates, analysis of isolates for penicillin-binding protein signature, determination of global pneumococcal sequence clusters, MLST alleles, serotypes, sequence types and acquired resistance genes (mefA, ermB, tetM, folA/P, cat), identification of virulence genes were carried out.
Results. Twenty-eight GPSCs, 45 sequence types and 27 serotypes were identified. The coverage rates of PPV-23 and PCV-13 were 78% and 59%, respectively. Serotypes 3 (18%), 19F (9%), 23F (7%) and 15B (6%) were predominant. The GPSC12 lineage (serotype 3) was predominant (43%). Lineages expressing vaccine serotypes GPSC1(19F), GPSC6(14), GPSC13(6A), GPSC904(14) and GPSC10(19F) exhibited multiple antimicrobial resistance, including penicillin resistance. The resistant lineages expressing non-vaccine serotypes were GPSC230 (13) and GPSC177 (35F). In most cases, genotypic and phenotypic resistance to penicillin (increased MICs of β-lactams correlated with types of penicillin-binding proteins), erythromycin (ermB, mefA, ermB/mefA), clindamycin (ermB) and tetracycline (tetM), and trimethoprim-sulfamethoxazole (folA, folP) was found to be consistent. The virulence genes cbpG, lytA, pce/cbpE, pavA, pfbA, ply, hysA, nanA and cps4A were detected in all isolates. Zinc metalloproteinase C was detected in 13% of isolates.
Conclusion. A high diversity of serotypes and lineages among pneumococcal isolates from meningitis patients was revealed. Out of the 68 S. pneumoniae isolates from patients with bacterial meningitis, more than 17% belonged to non-vaccine serotypes. The results of phenotypic and genotypic antimicrobial resistance comparison were characterized by good concordance, which indicates the necessity for further study of the possibility of using whole-genome sequencing as a diagnostic tool to identify resistance mechanisms in clinical isolates of pneumococci.
Full Text
Introduction
Streptococcus pneumoniae is a human respiratory pathogen and a major cause of morbidity and mortality worldwide. S. pneumoniae is the 4th most common cause of fatal infections, such as septicemia and meningitis, and is estimated by the World Health Organization (WHO) to cause 1.6 million deaths, of which 0.7–1.0 million occur in children under 5 years of age, mostly in developing countries [1–3]. Surface capsular polysaccharides of S. pneumoniae are one of the most important virulence factors and the basis of pneumococcal serotyping. Currently, more than 100 serotypes of S. pneumoniae are known [4]. Due to its ability to acquire exogenous DNA, pneumococcus can switch serotypes and acquire antibiotic resistance genes [5]. Uncontrolled use of antimicrobials, selective pressure of pneumococcal vaccines, high level of genetic recombination of S. pneumoniae inevitably lead to changes in pneumococcal population: emergence of new non-vaccine serotypes, emergence of isolates with multiple antimicrobial resistance, change in virulence profile. Due to the current situation in the world, in 2024 WHO included macrolide-resistant S. pneumoniae in the updated list of priority bacterial pathogens of intermediate level in the world1.
Currently, whole-genome sequencing technologies allow obtaining information on genetic changes, serotypes, sequencing types determined both by the classical 7-locus scheme and by the core genome MLST, virulence profile, antimicrobial resistance status of pneumococci, which is important for epidemiologic surveillance [6–10].
The aim of the study: to provide key characteristics of S. pneumoniae isolates from pneumococcal meningitis patients circulating in Russia in 2015–2020, based on high-throughput sequencing data, including global pneumococcal sequence clusters, serotypes, virulence factors and genetic determinants of resistance, in comparison with clinical data on susceptibility to antimicrobials.
Materials and methods
The study of virulence factors and resistance genes in 68 invasive isolates of S. pneumoniae isolated from blood and cerebrospinal fluid of patients diagnosed with bacterial meningitis was performed. All isolates were obtained during different stages of the PeGAS multicenter study in 2015–2020 in different regions of Russia [11]. The isolation and primary identification of isolates were performed in local microbiological laboratories of the centers participating in the study as part of the standard procedure for bacteriological examination of biological material obtained from patients diagnosed with bacterial meningitis and in accordance with MG 4.2.1887-04 “Laboratory diagnosis of meningococcal infection and purulent bacterial meningitis”. S. pneumoniae isolates were transported to the central laboratory of the Research Institute of Antimicrobial Chemotherapy (RIAC) on modified Dorset medium. The RIAC evaluated the compliance of the sent isolates with the inclusion criteria and performed their identification based on colony morphology on blood agar (NEM), the presence of α-hemolysis, negative catalase reaction, sensitivity to optoquine (Oxoid) and positive results of latex-agglutination using the DrySpot Pneumo kit (Oxoid). All isolates were stored in tubes with trypticase-soy broth (bioMerieux) supplemented with 30% sterile glycerol (Sigma) at –70°C until antimicrobial sensitivity was determined. Contaminated and non-viable isolates were excluded from the study. Sensitivity to antimicrobials was determined by broth microdilution in accordance with the requirements of ISO 20776-1:20202, and the sensitivity categories of isolates to antimicrobials were determined based on borderline values of minimum inhibitory concentrations (MIC) in accordance with EUCAST standards3 and Russian recommendations. To control the quality of sensitivity determination, a reference strain of S. pneumoniae ATCC 49619 was tested in parallel with the isolates under study.
Whole-genome sequencing was performed at the Central Research Institute of Epidemiology of Rospotrebnadzor. Sample preparation was performed using the Nextera (Illumina) protocol. High-throughput sequencing was performed on a HiSeq 1500 instrument using HiSeq PE Rapid Cluster Kit v2 and HiSeq Rapid SBS Kit v2 (Illumina). Whole-genome nucleotide sequences were assembled using the SPAdes v. 3.13 program. A detailed description of the sample preparation methodology for whole-genome sequencing was given earlier [12].
The whole genomic nucleotide sequences of the studied isolates, data on serotypes and antibiotic sensitivity, as well as information on the sources of strains were deposited in the PubMLST database4: accession numbers: 51080–51125, 73010–73011, 73013–73015, 73017–73033.
Invasive pneumococcal isolates were analyzed for the PBP signature, where the combination of 3 signatures (PBP1A, PBP2B, PBP2X) determines the level of resistance to β-lactams, global pneumococcal sequence clusters were identified, MLST alleles, serotypes, sequence types and acquired resistance genes (mefA, ermB, tetM, folA/P, cat) using a tool available on Pathogenwatch, a global genomic surveillance platform5. Virulence genes were identified using the online AMRseq program6 and the BacWGSTdb program7.
The study was non-interventional and did not involve comparison of groups. Descriptive statistics methods were used to present the results, with the determination of the absolute and relative number of observations.
Results
Molecular and biological characterization of invasive S. pneumoniae isolates
We identified 28 global pneumococcal sequence clusters (GPSCs), 45 sequence types and 27 serotypes (Table 1).
Table 1. Global clusters of pneumococcal sequences associated with sequence types and serotypes
GPSC | Sequence types | Serotype | Number of |
1 | 236 | 19F | 2 (7) |
2 | 15249 | 1 | 1 (4) |
3 | 1012 | 11A | 1 (4) |
6 | 3418, 143 | 6E(6B),14 | 2 (7) |
7 | 311, 152248, 16095, 311 | 23F | 4 (14) |
10 | 230 | 19F | 1 (4) |
11 | 1262 | 15B | 2 (7) |
12 | 505, 180, 15251, 15250, 2049 | 3 | 12 (43) |
13 | 12493, 473 | 6A, 6B | 2 (7) |
16 | 66, 16098 | 9N | 2 (7) |
19 | 433 | 22F | 2 (7) |
32 | 3244, 11901,2824, 3544 | 7F, 8 | 4 (14) |
38 | 393 | 38 | 1 (4) |
43 | 239 | 9V | 3 (11) |
44 | 179 | 19F | 1 (4) |
49 | 440 | 23F | 1 (4) |
68 | 15252, 3187 | 18C | 2 (7) |
76 | 490 | 6A | 1 (4) |
98 | 1480 | 8 | 1 (4) |
123 | 447 | 37 | 1 (4) |
162 | 2361 | 4 | 2 (7) |
177 | 2991 | 35F | 2 (7) |
212 | 6202 | 12F, 15F | 4 (14) |
229 | 1025 | 15B, 15C | 3 (11) |
230 | 2754 | 13 | 1 (4) |
365 | 225 | 28A | 2 (7) |
376 | 9247 | 6E(6B) | 1 (4) |
904 | 782 | 14 | 1 (4) |
Not assigned | 15 | 19F | 2 (7) |
Not assigned | 15247 | 10С | 1 (4) |
Not assigned | 5205 | 8 | 1 (4) |
Not assigned | 13459 | 10А | 1 (4) |
Not assigned | 16099 | 4 | 1 (4) |
Note. Not assigned — global pneumococcal sequence cluster number is not assigned.
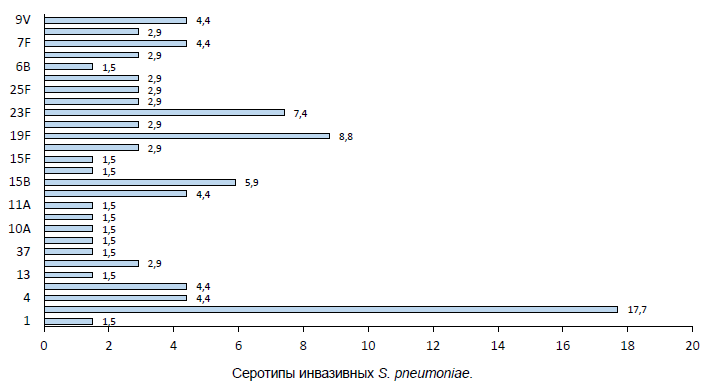
Analysis of whole genomic data showed that serotypes included in pneumococcal polysaccharide 23-valent vaccine (PPV-23) and pneumococcal conjugate vaccine 13 (PCV-13) were predominant among invasive isolates — 3 (18%), 19F (9%), 23F (7%) and 15B (6%). The coverage rate of PPV-23 was 79% and PCV-13 was 59%. Frequency of isolation of pneumococci of non-vaccine serotypes in meningitis in Russia: 28A and 35F — 3% each, 13, 37, 38, 10C, 15C and 15F — more than 1% (Figure).
Serotypes of invasive S. pneumoniae.
Of the 28 GPSCs, 20 GPSCs were of vaccine serotypes and 6 GPSCs were of non-vaccine serotypes. Two lineages, GPSC212 (12F, 15F) and GPSC229 (15B, 15C) expressed both vaccine and non-vaccine serotypes. The GPSC12 lineage expressed only vaccine serotype 3 and accounted for more than 42%. In some cases, the same serotype was associated with different lineages. The second most frequent serotype 19F was expressed by lineages GPSC1, 10, 44 and a new lineage whose GPSC is undefined, serotype 23F by GPSC7 and GPSC49, and serotype 15B by GPSC11 and GPSC229. Non-vaccine serotypes were expressed by the lineages GPSC 365 (serotype 28A), GPSC177 (serotype 35F), GPSC212 (serotype 15F), GPSC229 (serotype 15C), GPSC123 (serotype 37), GPSC38 (serotype 38), and GPSC230 (serotype 13).
Antimicrobial sensitivity of invasive isolates of S. pneumoniae to antimicrobials
Analysis of sensitivity to antimicrobials showed that 6 (9%) invasive isolates were resistant to penicillin (Table 2), 11 (16%) to tetracycline, 5 (7%) to erythromycin, 2 (3%) to clindamycin, and 1 (1%) to respiratory fluoroquinolones. To trimethoprim-sulfamethoxazole 12 (18%) isolates were resistant and 12 (18%) were sensitive at increased exposure. Among the isolates of 28 GPSC lineages, isolates of 5 lineages expressing vaccine serotypes were resistant to 3 or more classes of antimicrobials simultaneously: GPSC1 (serotype 19F), GPSC6 (serotype 14) were resistant to penicillin, tetracycline, trimethoprim-sulfamethoxazole, erythromycin, and clindamycin; GPSC10 (serotype 19F), GPSC904;9 (serotype 14) — to penicillin, tetracycline and trimethoprim-sulfamethoxazole, GPSC6 (serotype 6E(B)) — to penicillin, tetracycline and trimethoprim-sulfamethoxazole, GPSC6 (serotype 6E(B)) — to penicillin, erythromycin, and trimethoprim-sulfamethoxazole. One lineage with vaccine serotypes was resistant to two classes of antibiotics simultaneously: GPSC44 (serotype 19F) to tetracycline and erythromycin. Two lineages expressing non-vaccine serotypes, GPSC177 (serotype 35F) and GPSC230 (serotype 13), were resistant to tetracycline and trimethoprim-sulfamethoxazole.
Genetic determinants of antimicrobial resistance
In clinical isolates of S. pneumoniae, resistance to β-lactams is primarily due to variations in amino acid sequences in the transpeptidase domains of penicillin-binding proteins (PBPs): PBP1a, PBP2b and PBP2x, which reduce the affinity of β-lactam antibiotics for these sites. The type of PBPs can predict the levels of resistance to β-lactams [13]. Analysis of the results of whole-genome sequencing of invasive S. pneumoniae isolates revealed that the most common type of PBP in sensitive isolates (PBP1a-PBP2b-PBP2x) was 2-0-2 — 29%, 11-4-0 — 4% and 12-0-6 — 3% were less common. All penicillin-resistant isolates had signatures: 13-16-47, 17-15-22, 24-53-77, 36-34-44, and 31-12-18. Several combinations of new PBP types were also identified: new-27-new, 34-11-new. Resistance to macrolides end lincosamides (erythromycin and clindamycin) was due to the presence of the methylase (ermB) gene in 5 (7%) isolates, while 1 (1%) isolate had both ermB and macrolide efflux pump (mefA/E) genes detected simultaneously. Resistance to trimethoprim-sulfamethoxazole was associated with substitution in folA I100L and/or insertion of 1 or 2 foP codons, whereas isolates categorized as sensitive at higher exposure often had insertion of 1 or 2 codons in folP. Chloramphenicol resistance was predicted by the presence of the chloramphenicol acetyltransferase gene (cat), while fluoroquinolone resistance was predicted by mutations in gyrA and parC genes.
Comparison of resistance phenotype and genotype
The concordance of resistance genotype and phenotype was generally high, but in some cases there were discrepancies between genotypic and phenotypic resistance.
Resistance to β-lactams showed good concordance. Six penicillin-resistant pneumococcal isolates had PBP signatures characteristic of resistant pneumococci. Interestingly, all 6 penicillin-resistant isolates had PBP1a(13-36)-PBP2b(12-53)-PBP2x(18-77) signatures, whereas all sensitive isolates had low-numbered PBP signatures. Thus, PBP transpeptidase signatures are reliable indicators of the MICs of various β-lactam antibiotics in clinical isolates of pneumococci and can serve as an alternative to phenotypic sensitivity testing.
All 11 tetracycline-resistant pneumococci carried tetM genes. Three isolates containing tetM were categorized as sensitive, which may be due to mutations in tetM not considered in our study.
Table 2. Resistance of pneumococcal lineages GPSC to antimicrobials
GPSC | Serotype | Number | Genotype, resistance, n (%) | ||||||
PEN | TET | TS | CL | ERY | CHL | FX | |||
1 | 19F | 2 | 2 (3) | 2 (3) | 2 (3) | 1 (1) | 2 (3) | 0 | 0 |
2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
3 | 11A | 1 | 0 | 0 | 1 (1) | 0 | 0 | 0 | 0 |
6 | 6E(B),14 | 2 | 1 (1) | 2 (3) | 2 (3) | 2 (3) | 2 (3) | 0 | 0 |
7 | 23F | 4 | 0 | 0 | 4 (6) | 0 | 0 | 0 | 0 |
10 | 19F | 1 | 1 (1) | 1 (1) | 1 (1) | 0 | 0 | 0 | 0 |
11 | 15B | 2 | 0 | 0 | 2 (3) | 0 | 0 | 0 | 0 |
12 | 3 | 12 | 0 | 1 (1) | 0 | 0 | 0 | 0 | 1 (1) |
13 | 6A, 6B | 2 | 1 (1) | 1 (1) | 2 (3) | 0 | 1 (1) | 0 | 0 |
16 | 9N | 2 | 0 | 0 | 1 (4) | 0 | 0 | 0 | 1 (1) |
19 | 22F | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
32 | 7F,8 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
38 | 38 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
43 | 9V | 3 | 0 | 0 | 3 (4) | 0 | 0 | 0 | 0 |
44 | 19F | 1 | 0 | 1 (1) | 1 (1) | 1 (1) | 1 (1) | 0 | 0 |
49 | 23F | 1 | 0 | 0 | 1 (1) | 0 | 0 | 0 | 0 |
68 | 18C | 2 | 0 | 0 | 2 (3) | 0 | 0 | 0 | 0 |
76 | 6A | 1 | 0 | 0 | 1 (1) | 0 | 0 | 0 | 0 |
98 | 8 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
123 | 37 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
162 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
177 | 35F | 2 | 0 | 2 (3) | 2 (3) | 0 | 0 | 0 | 0 |
212 | 12F, 15F | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
229 | 15B, 15C | 3 | 0 | 0 | 3 (4) | 0 | 0 | 0 | 0 |
230 | 13 | 1 | 0 | 1 (1) | 1 (1) | 0 | 1 (1) | 0 | 0 |
365 | 28A | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
376 | 6E(6B) | 1 | 0 | 0 | 1 (1) | 0 | 0 | 1 (4) | 0 |
904;9 | 14 | 1 | 1 (1) | 1 (1) | 1 (1) | 0 | 0 | 0 | 0 |
Not assigned | 10С, 19F, 8, 10A ,4 | 6 | 0 | 2 (3) | 2 (3) | 0 | 0 | 0 | 0 |
28 | 27 | 68 | 6 (9) | 14 (21) | 33 (49) | 4 (6) | 7 (10) | 1 (1) | 2 (3) |
Note. PEN, penicillin resistance predicted based on PBP1a, PBP2b and PBP2x sequences; TET, tetracycline resistance predicted by the presence of the tet M gene; TS, trimethoprim-sulfamethoxazole resistance, associated with substitution in folA I100L and/or insertion of 1 or 2 codons in folP_aa_insert_57-70, CL — clindamycin resistance predicted of gene erm B; ERY — macrolide resistance predicted by the presence of the methylase gene (ermB) and macrolide efflux pump gene (mefA/E); CHL — chloramphenicol resistance predicted by the presence of the chloramphenicol acetyltransferase gene (cat); FX — fluoroquinolone resistance predicted by the presence of mutations in the gyrA, parC genes.
In the case of trimethoprim-sulfamethoxazole, a good correlation between the presence of resistance markers and phenotypic resistance was found for most isolates. Twelve isolates were double mutants with a substitution in folA I100L and an insertion of 1 or 2 codons in folP_aa_insert_57-70 and were resistant to trimethoprim-sulfamethoxazole (Table 2). Twelve pneumococcal isolates containing insertions in folP had an IPC of 2 mg/L and were categorized as sensitive at increased exposure, and 9 isolates with single mutations in folP were categorized as sensitive (IPC ≤ 1 mg/L).
All 5 erythromycin-resistant isolates carried the ermB gene, 1 resistant isolate carried both ermB and mefA, while 2 isolates with the mefA gene remained phenotypically sensitive to erythromycin. The 2 clindamycin-resistant isolates carried ermB genes, but ermB was also detected in 2 phenotypically sensitive isolates.
Due to the fact that there are no criteria for determining the category of sensitivity to chloramphenicol for S. pneumoniae, the activity of this drug was assessed based on the epidemiological cut-off value. The MIC of chloramphenicol for all isolates tested was less than 8 mg/L, which corresponds to the wild-type population, but 1 isolate carried cat gene.
One isolate was found to be resistant to respiratory fluoroquinolones (levofloxacin and moxifloxacin); it had mutations in the gyrA and parC genes. At the same time, a mutation in the parC gene was detected in 1 isolate among fluoroquinolone-sensitive pneumococci.
Genetic determinants of pneumococcal virulence
To gain insight into the genetic features contributing to virulence, we examined the presence of the major protein virulence factors of pneumococcus. Choline-binding proteins (CbpG, LytA and Pce/CbpE), PavA and PfbA, known as fibronectin- and plasminogen-binding proteins, as well as hyaluronidase, pneumolysin, neuraminidase, and capsule-associated Cps4A were detected in all invasive pneumococcal isolates (Table 3). Zinc metalloproteinase C was detected in 9 (13%) isolates.
Table 3. Characteristics of virulence genes
Virulence gene | Name of the encoded protein | % idеntity | Number of isolates | |
n | % | |||
cbpG | Choline-binding protein G | 99,30 | 68 | 100 |
lytA | Autolysine | 98,75 | 68 | 100 |
pce/cbpE | Choline-binding protein E | 99,18 | 68 | 100 |
ply | Pneumolysine | 99,86 | 68 | 100 |
pavA | Fibronectin-binding protein | 99,52 | 68 | 100 |
pfbA | Plasmin and fibronectin-binding protein A | 99,72 | 68 | 100 |
hysA | Hyaluronidase | 99,16 | 68 | 100 |
nanA | Neuraminidase A | 98,77 | 68 | 100 |
cps4A | Capsule synthesis | 96,54 | 68 | 100 |
zmpC | Zinc metalloproteinase C | 99,96 | 9 | 13 |
Discussion
In our study, pneumococci of serotype 3 prevailed among clinical isolates causing meningitis in Russia. A similar situation is observed in many other countries during the postvaccination period. In Austria, England, Canada, Sweden and Germany, a significant increase in invasive pneumococcal diseases of serotype 3 in adults has been observed over the last 3 years [14]. In Brazil, serotype 3 became the predominant cause of invasive disease in the post-PCV era among adults [15, 16]. The low efficacy of conjugate vaccines against serotype 3 pneumococci is related to the structure of the polysaccharide capsule, which is non-covalently bound to cell wall peptidoglycan [17–21]. It should be noted that isolates of the GPSC12 lineage (serotype 3), remaining the main cause of invasive forms of pneumococcal infection worldwide after the introduction of PCV13, usually retain sensitivity to antimicrobials [22–25]. The low incidence of antimicrobial resistance in serotype 3 isolates may be associated with the high invasiveness of this serotype and the relatively short duration of carriage, which, in turn, reduces the impact of antimicrobials in the treatment of infections of other etiologies [26]. At the same time, a serotype 3 study conducted in England [27] revealed that since 2018, GPSC12 lineage isolates resistant to penicillin, macrolides, chloramphenicol and tetracycline have been emerging [28, 29]. The increasing resistance of serotype 3 isolates indicates the circulation of more antibiotic-resistant clones [30]. In our study, all S. pneumoniae serotype 3 isolates were sensitive to penicillin; of 12 isolates isolated from cerebrospinal fluid, only 2 contained resistance genes to fluoroquinolones — parC and to tetracycline — tetM, while retaining phenotypic sensitivity to these drugs.
Pneumococci of serotype 19F were the second most frequently isolated in meningitis, which may be due to the peculiarities of the capsular polysaccharide, which is more resistant to the deposition of the C3b component of complement and antibodies on the bacterial walls, which reduces the sensitivity to opsonophagocytosis [31, 32]. Our results indicate that the circulation of serotype 19F is associated with the spread of 3 lineages: GPSC10, GPSC1 and GPSC44. All lineages were characterized by resistance to various antimicrobials, with the GPSC10 and GPSC1 lineages showing multiple antimicrobial resistance. In Canada, 19F serotype dominance was associated with the spread of GPSC1, GPSC4, GPSC9, GPSC10, GPSC18, GPSC44 and GPSC119 lineages [33], in Sweden with the GPSC1 lineage [34], and in Asia, Europe, North America, and South America, 19F serotype was one of the dominant serotypes in the GPSC1 lineage [35]. In South Africa, the dominance of the 19F serotype was associated with the spread of the GPSC1 and GPSC21 lineages, and a high, about 50%, hospital-acquired mortality rate from pneumococcal meningitis was found to be associated with the 19F serotype [35, 36]. The increased incidence of 19F serotype after the introduction of PCV13 in India has been associated with the multidrug-resistant GPSC1 and GPSC10 lineages, with GPSC10 being of particular note as it expressed both vaccine serotypes, including 19F, and non-vaccine serotypes, and thus contributed most to the spread of non-vaccine serotypes among clinical isolates [37]. The GPSC10 lineage is capable of simultaneously expressing a wide range of serotypes, which facilitates its adaptation to selective vaccine pressure. An international dataset of the GPSC10 lineage showed that this lineage expresses 16 serotypes, of which only 6 are included in PCV13. Moreover, the GPSC10 lineage has a relatively high potential to develop invasive forms of infection and a propensity to cause meningitis, regardless of serotype [38]. It was found that it took about 3-5 years for pneumococci of the GPSC10 lineage to spread in France, and in Spain, Argentina, and Israel a rapid change in the serotype composition of this lineage occurred in the post-vaccination period. Thus, together with its transmissibility, GPSC10 should be considered as a high-risk lineage that may eventually reduce the efficacy of vaccines worldwide [39, 40]. In a study conducted by E. Egorova et al. found that in Russia in 2011-2018, serotype 19F isolates belonged to 8 different lineages (GPSC1, GPSC44, GPSC10 GPSC6, GPSC11, GPSC18, GPSC43 and GPSC591), of which the lineages GPSC1, GPSC6 and GPSC10 were characterized by resistance to antimicrobials [41].
According to the results of the present study, pneumococci of serotype 23F were the third most common serotype responsible for the development of bacterial meningitis in Russia. Four out of 5 isolates of serotype 23F belonged to the GPSC7 lineage, 1 — to the GPSC49 lineage. All isolates of serotype 23F were characterized by resistance to trimethoprim-sulfamethoxazole. In studies conducted in China and Iran, serotype 23F was one of the dominant serotypes isolated from the cerebrospinal fluid of patients [42, 43]. The results of studies performed in the UK showed that serotype 23F isolates were part of the GPSC7 lineage. While in 2002 this lineage was dominated by serotype 23F and had only a small number of serotype 23A, in 2009 isolates of serotype 23B appeared [29]. The GPSC7 lineage was one of the dominant lineages responsible for invasive disease during the PCV13 era in Hong Kong, Israel, Malawi, South Africa, Gambia and the USA [22].
The results of international studies indicate that the post-vaccination period is characterized by a decrease in the proportion of vaccine serotypes and an increase in non-vaccine pneumococcal serotypes among different age groups of the population [44, 45]. Lineages represented by vaccine serotypes in which non-vaccine serotypes were already present persist in the population [15]. It has been observed that these non-vaccine serotypes have a high potential to develop invasive forms of infection [46, 47]. The results of our study indicate that in Russia more than 17% of pneumococcal meningitis cases were caused by pneumococci of non-vaccine serotypes. The relatively high proportion of isolates with serotype 15B (6%) and the presence of isolates with non-vaccine serotypes 28A, 37 and 38, which were not previously associated with pneumococcal meningitis in Russia, are noteworthy. The level of antibiotic resistance was found to be lower in non-vaccine serotypes than in vaccine serotypes and differed depending on the GPSC lineage. In our study, lineages expressing non-vaccine serotypes were sensitive to all tested antimicrobials, except for 2 lineages (Table 2).
Different virulence factors are involved in the process of invasive infection at different stages. According to the results of our study, classical genes encoding virulence factors such as capsule synthesis, pneumococcal surface adhesin, autolysin, fibronectin binding protein and pneumolysin were found in all invasive isolates. An interesting observation was made regarding the zmpC gene. It was detected only in 9 invasive pneumococcal isolates of the pneumococcal lines GSPC229 (serotype 15C,15B), GSPC3 (serotype 11A), GSPC162 (serotype 4), GSCP904 (serotype 14) and the new line GSPC (serotype 8). Previously, in a study in the Netherlands, the zmpC gene was identified in invasive serotypes 8, 11A and 4 belonging to the GPSC3 lineage. It was found that cases of invasive pneumococcal infections caused by zmpC-positive pneumococci were more often accompanied by sepsis [48]. Japanese scientists suggested that the zinc metalloprotease zmpC suppresses the virulence of pneumococci by inhibiting bacterial invasion into the central nervous system [49]. Thus, zinc metalloprotease zmpC is of particular interest and requires additional research.
Conclusion
- 28 global pneumococcal sequence clusters (GPSC) and 45 S. pneumoniae sequence-types associated with invasive strains were identified in Russia. Of 68 S. pneumoniae isolates from patients with bacterial meningitis, more than 17% belonged to non-vaccine serotypes.
- Antibiotic resistance of pneumococci of vaccine serotypes was higher than that of non-vaccine serotypes.
- The emergence of non-vaccine serotype lineages of pneumococcus with determinants of high virulence, including resistance to antibiotics, necessitates further research into the molecular genetic characterization of isolates causing meningitis.
- The results comparing phenotypic and genotypic antimicrobials were characterized by good concordance, indicating the need to further explore the possibility of using whole-genome sequencing as a diagnostic tool to identify resistance mechanisms in clinical isolates of S. pneumoniae.
In conclusion, the characterization of pneumococcal lineages and their genetic variations that influence resistance and invasiveness are highly informative for establishing a global strategy for continuous epidemiological surveillance of the pneumococcal population.
1 WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance.
URL: https://who.int/publications/i/item/9789240093461
2 ISO 207761:2019 Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices. Part 1: Broth microdilution reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases.
3 European Committee on Antimicrobial Susceptibility testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Ver. 14.0, 2024. URL: www. eucast.org/clinical_breakpoints/ (data of access: 01.11.2024).
4 Streptococcus pneumoniae MLST Databases.
URL: https://pubmlst.org/spneumoniae/ (data of access: 17.02.2020).
5 A Global Platform for Genomic Surveillancе.
URL: https://pathogen.watch
6 AMRseq. URL: https://amrseq.net/ru/
7 BacWGSTdb. URL: http://bacdb.cn/BacWGSTdb/index.php
About the authors
Aida N. Chagaryan
Research Institute of Antimicrobial Chemotherapy, Smolensk State Medical University
Author for correspondence.
Email: aida.chagaryan@antibiotic.ru
ORCID iD: 0000-0001-9195-8764
Cand. Sci. (Biol.), researcher, Laboratory of antibiotic resistance, Research Institute of Antimicrobial Chemotherapy
Россия, SmolenskNatali V. Ivanchik
Research Institute of Antimicrobial Chemotherapy, Smolensk State Medical University
Email: natali.ivanchik@antibiotic.ru
ORCID iD: 0000-0002-9392-0732
Cand. Sci. (Med.), researcher, Laboratory of antibiotic resistance, Research Institute of Antimicrobial Chemotherapy
Россия, SmolenskAlexey Yu. Kuzmenkov
Research Institute of Antimicrobial Chemotherapy, Smolensk State Medical University
Email: alexey.kuzmenkov@antibiotic.ru
ORCID iD: 0000-0001-9562-2096
Sci. (Med.), Professor, Deputy director, Associate Professor, Microbiology department, Institute of Antimicrobial Chemotherapy
Россия, SmolenskRoman S. Kozlov
Research Institute of Antimicrobial Chemotherapy, Smolensk State Medical University; Central Research Institute of Epidemiology
Email: roman.kozlov@antibiotic.ru
ORCID iD: 0000-0001-8728-1113
D. (Med.), Professor, Director, Research Institute of Antimicrobial Chemotherapy, Chancellor
Россия, Smolensk; MoscowIrina I. Gaponova
Central Research Institute of Epidemiology
Email: gaponova@cmd.su
ORCID iD: 0000-0003-4481-2249
research laboratory assistant, Scientific group for the development of new methods for detecting genetic polymorphisms
Россия, MoscowKonstantin O. Mironov
Central Research Institute of Epidemiology
Email: mironov@pcr.ru
ORCID iD: 0000-0001-8207-9215
Sci. (Med.), Head, Group for the development of new methods for detecting genetic polymorphisms
Россия, MoscowReferences
- Principi N., Di Cara G., Bizzarri I., et al. Prevention of invasive pneumococcal disease: problems emerged after some years of the 13-valent pneumococcal conjugate vaccine use. Curr. Infect. Dis. Rep. 2018;20(1):1. DOI: https://doi.org/10.1007/s11908-018-0607-z
- Briles D.E., Paton J.C., Mukerji R., et al. Pneumococcal vaccines. Microbiol. Spectr. 2019;7(6):10.1128/microbiolspec.gpp3-0028-2018. DOI: https://doi.org/10.1128/microbiolspec.GPP3-0028-2018
- Chen H., Matsumoto H., Horita N., et al. Prognostic factors for mortality in invasive pneumococcal disease in adults: a systematic review and meta-analysis. Sci. Rep. 2021;11(1):11865. DOI: https://doi.org/10.1038/s41598-021-91234-y
- Ganaie F., Saad JS., McGee L., et al. A new pneumococcal capsule type, 10D, is the 100th serotype and has a large cps fragment from an oral streptococcus. mBio. 2020;11(3):e00937-20. DOI: https://doi.org/10.1128/mbio.00937-20
- Salvadori G., Junges R., Morrison D.A., Petersen F.C. Competence in Streptococcus pneumoniae and close commensal relatives: mechanisms and implications. Front. Cell. Infect. Microbiol. 2019;9:94. DOI: https://doi.org/10.3389/fcimb.2019.00094
- Протасова И.Н., Бахарева Н.В., Перьянова О.В. и др. Молекулярно-эпидемиологическая характеристика и резистентность пневмококков у детей дошкольного возраста. Сибирское медицинское обозрение. 2018;(3):73–9. Protasova I.N., Bakhareva N.V., Peryanova O.V., et al. Molecular-epidemiological characteristics and resistance of pneumococcus in children of preschool age. Siberian Medical Review. 2018;(3):73–9. DOI: https://doi.org/10.20333/2500136-2018-3-73-79 EDN: https://elibrary.ru/xserud
- Sidorenko S., Rennert W., Lobzin Y., et al. Multicenter study of serotype distribution of Streptococcus pneumoniae nasopharyngeal isolates from healthy children in the Russian Federation after introduction of PCV13 into the National Vaccination Calendar. Diagn. Microbiol. Infect. Dis. 2020;96(1):114914. DOI: https://doi.org/10.1016/j.diagmicrobio.2019.114914
- Протасова И.Н., Мартынова Г.П., Ильенкова Н.А. и др. Этиологическая роль и молекулярно-генетические особенности Streptococcus pneumoniae при инфекционных заболеваниях у детей. Детские инфекции. 2020;19(1):7–12. DOI: https://doi.org/10.22627/2072-8107-2020-19-1-7-12 EDN: https://elibrary.ru/dwibms
- Миронов К.О., Гапонова И.И., Корчагин В.И. и др. Антигенная и генетическая характеристика штаммов Streptococcus pneumoniae, выделенных от больных инвазивными и неинвазивными пневмококковыми инфекциями, с использованием высокопроизводительного секвенирования. Журнал микробиологии, эпидемиологии и иммунобиологии. 2021;98(5):512–8. Mironov K.O., Gaponova I.I., Korchagin V.I., et al. Antigenic and genetic characterization of Streptococcus pneumoniae strains isolated from patients with invasive and non-invasive pneumococcal infections by using high-throughput sequencing. Journal of Microbiology, Epidemiology and Immunobiology. 2021;98(5):512–8. DOI: https://doi.org/10.36233/0372-9311-144, EDN: https://elibrary.ru/kvjhkq
- Vohrnová S., Kozáková J. Posibilities for use of whole genome sequencing (WGS) for the analysis of Streptococcus pneumoniae isolates. Epidemiol. Mikrobiol. Imunol. 2024;73(1):30–6. DOI: https://doi.org/10.61568/emi/11-6254/20240123/136240 (in Czech)
- Иванчик Н.В., Чагарян А.Н., Сухорукова М.В. и др. Антибиотикорезистентность клинических штаммов Streptococcus pneumoniae в России: результаты многоцентрового эпидемиологического исследования «ПеГАС 2014–2017». Клиническая микробиология и антимикробная химиотерапия. 2019;21(3):230–7. Ivanchik N.V., Chagaryan A.N., Sukhorukova M.V., et al. Antimicrobial resistance of clinical Streptococcus pneumoniae isolates in Russia: the results of multicenter epidemiological study «Pehasus 2014–2017». Clinical Microbiology and Antimicrobial Chemotherapy. 2019;21(3):230–7. DOI: https://doi.org/10.36488/cmac.2019.3.230-237, EDN: https://elibrary.ru/hlaxrl
- Миронов К.О., Корчагин В.И., Михайлова Ю.В. и др. Характеристика штаммов Streptococcus рneumoniae, выделенных от больных инвазивными пневмококковыми инфекциями, с использованием высокопроизводительного секвенирования. Журнал микробиологии, эпидемиологии и иммунобиологии. 2020;97(2):113–8. Mironov K.O., Korchagin V.I., Mikhailova Yu.V., et al. Characterization of Streptococcus pneumoniae strains causing invasive infections using whole-genome sequencing. Journal of Microbiology, Epidemiology and Immunobiology. 2020;97(2):113–8. DOI: https://doi.org/10.36233/0372-9311-2020-97-2-113-118 EDN: https://elibrary.ru/lnxmqy
- Kawaguchiya M., Urushibara N., Aung M.S., et al. Genetic characterization of penicillin-binding proteins of nonencapsulated Streptococcus pneumoniae in the postpneumococcal conjugate vaccine era in Japan. Int. J. Infect. Dis. 2022;120:174–6. DOI: https://doi.org/10.1016/j.ijid.2022.04.033
- Grant L.R., Slack M.P.E., Theilacker C., et al. Distribution of serotypes causing invasive pneumococcal disease in older adults from high-income countries and impact of pediatric and adult vaccination policies. Vaccine. 2023;41(38):5662–9. DOI: https://doi.org/10.1016/j.vaccine.2023.08.001
- Brandileone M.C.C., Almeida S.C.G., Minamisava R., Andrade A.L. Distribution of invasive Streptococcus pneumoniae serotypes before and 5 years after the introduction of a 10-valent pneumococcal conjugate vaccine in Brazil. Vaccine. 2018;36(19):2559–66. DOI: https://doi.org/10.1016/j.vaccine.2018.04.010
- Almeida S.C.G., Cassiolato A.P., Dias U.J., et al. Molecular characterization of invasive Streptococcus pneumoniae isolated in pre (2005–2009) and post (2011–2015) 10-valent pneumococcal conjugate vaccine introduction in Brazil. In: 37th Annual Meeting of the European Society for Paediatric Infectious Diseases. Ljubljana, Slovenia;2019.
- Shenoy A.T., Beno S.M., Brissac T., et al. Severity and properties of cardiac damage caused by Streptococcus pneumoniae are strain dependent. PLoS One. 2018;13(9):e0204032. DOI: https://doi.org/10.1371/journal.pone.0204032
- Andrews N., Kent A., Amin-Chowdhury Z., et al. Effectiveness of the seven-valent and thirteen-valent pneumococcal conjugate vaccines in England: The indirect cohort design, 2006–2018. Vaccine. 2019;37(32):4491–8. DOI: https://doi.org/10.1016/j.vaccine.2019.06.071
- Wijayasri S., Hillier K., Lim GH., et al. The shifting epidemiology and serotype distribution of invasive pneumococcal disease in Ontario, Canada, 2007–2017. PLoS One. 2019;14(12):e0226353. DOI: https://doi.org/10.1371/journal.pone.0226353
- Goettler D., Streng A., Kemmling D., et al. Increase in Streptococcus pneumoniae serotype 3 associated parapneumonic pleural effusion/empyema after the introduction of PCV13 in Germany. Vaccine. 2020;38(3):570–7. DOI: https://doi.org/10.1016/j.vaccine.2019.10.056
- Babb R., Doyle C.R., Pirofski L.A. Isolation and characterization of human monoclonal antibodies to pneumococcal capsular polysaccharide 3. Microbiol. Spectr. 2021;9(3):e0144621. DOI: https://doi.org/10.1128/Spectrum.01446-21
- Lo SW., Gladstone RA., van Tonder AJ., et al. Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect. Dis. 2019;19(7):759–69. DOI: https://doi.org/10.1016/s1473-3099(19)30297-x
- Kandasamy R., Voysey M., Collins S., et al. Persistent circulation of vaccine serotypes and serotype replacement after 5 years of infant immunization with 13-valent pneumococcal conjugate vaccine in the United Kingdom. J. Infect. Dis. 2020;221(8):1361–70. DOI: https://doi.org/10.1093/infdis/jiz178
- Løchen A., Croucher N.J., Anderson R.M. Divergent serotype replacement trends and increasing diversity in pneumococcal disease in high-income settings reduce the benefit of expanding vaccine valency. Sci. Rep. 2020;10(1):18977. DOI: https://doi.org/10.1038/s41598-020-75691
- Kwun M.J., Ion A.V., Cheng H.C., et al. Post-vaccine epidemiology of serotype 3 pneumococci identifies transformation inhibition through prophage-driven alteration of a non-coding RNA. Genome Med. 2022;14(1):144. DOI: https://doi.org/10.1186/s13073-022-01147-2
- Butić I., Gužvinec M., Jelić M., et al. Serotype distribution and antimicrobial resistance of invasive Streptococcus pneumoniae isolates among Croatian adults during a fifteen-year period (2005–2019). Croat. Med. J. 2022;63(2):156–65. DOI: https://doi.org/10.3325/cmj.2022.63.156
- Azarian T., Mitchell P.K., Georgieva M., et al. Global emergence and population dynamics of divergent serotype 3 CC180 pneumococci. PLoS Pathog. 2018;14(11):e1007438. DOI: https://doi.org/10.1371/journal.ppat.1007438
- Sheppard C.L., Groves N., Andrews N., et al. The genomics of Streptococcus рneumoniae carriage isolates from UK children and their household contacts, pre-PCV7 to post-PCV13. Genes (Basel). 2019;10(9):687. DOI: https://doi.org/10.3390/genes10090687
- Groves N., Sheppard CL., Litt D., et al. Evolution of Streptococcus pneumoniae serotype 3 in England and Wales: a major vaccine evader. Genes (Basel). 2019;10(11):845. DOI: https://doi.org/10.3390/genes10110845
- Suaya J.A., Mendes R.E., Sings R.E., et al. Streptococcus pneumoniae serotype distribution and antimicrobial nonsusceptibility trends among adults with pneumonia in the United States, 2009‒2017. J. Infect. 2020;81(4):557–66. DOI: https://doi.org/10.1016/j.jinf.2020.07.035
- Sanapala S.R., Seco B.M.S., Baek J.Y., et al. Chimeric oligosaccharide conjugate induces opsonic antibodies against Streptococcus pneumoniae serotypes 19A and 19F. Chem. Sci. 2020;11(28):7401–7. DOI: https://doi.org/10.1039/d0sc02230f
- Downs SL., Olwagen CP., Van Der Merwe L., et al. Streptococcus pneumoniae and other bacterial nasopharyngeal colonization seven years post-introduction of 13-valent pneumococcal conjugate vaccine in South African children. Int. J. Infect. Dis. 2023;134: 45–52. DOI: https://doi.org/10.1016/j.ijid.2023.05.016
- Golden A.R., Adam H.J., Baxter M., et al. Whole genome characterization of Streptococcus pneumoniae from respiratory and blood cultures collected from Canadian hospitals before and after PCV-13 implementation in Canada: Focus on serotypes 22F and 33F from CANWARD 2007–2018. Vaccine. 2021;39(39): 5474–83. DOI: https://doi.org/10.1016/j.vaccine.2021.08.061
- Yamba Yamba L., Uddén F., Fuursted K., et al. Extensive/multidrug-resistant pneumococci detected in clinical respiratory tract samples in Southern Sweden are closely related to international multidrug-resistant lineages. Front. Cell. Infect. Microbiol. 2022;12:824449. DOI: https://doi.org/10.3389/fcimb.2022.824449
- Gladstone R.A., Lo S.W., Lees J.A., et al. International genomic definition of pneumococcal lineages, to contextualise disease, antibiotic resistance and vaccine impact. EBioMedicine. 2019;43:338–46. DOI: https://doi.org/10.1016/j.ebiom.2019.04.021
- Lekhuleni C., Ndlangisa K., Gladstone R.A., et al. Impact of pneumococcal conjugate vaccines on invasive pneumococcal disease-causing lineages among South African children. Nat. Commun. 2024;15(1):8401. DOI: https://doi.org/10.1038/s41467-024-52459-3
- Nagaraj G., Govindan V., Ganaie F., et al. Streptococcus pneumoniae genomic datasets from an Indian population describing pre-vaccine evolutionary epidemiology using a whole genome sequencing approach. Microb. Genom. 2021;7(9):000645. DOI: https://doi.org/10.1099/mgen.0.000645
- Lo S.W., Gladstone R.A., van Tonder A.J., et al. A mosaic tetracycline resistance gene tet(S/M) detected in an MDR pneumococcal CC230 lineage that underwent capsular switching in South Africa. J. Antimicrob. Chemother. 2020;75(3):512–20. DOI: https://doi.org/10.1093/jac/dkz477
- Gagetti P., Lo SW., Hawkins PA., et al. Population genetic structure, serotype distribution and antibiotic resistance of Streptococcus pneumoniae causing invasive disease in children in Argentina. Microb. Genom. 2021;7(9):000636. DOI: https://doi.org/10.1099/mgen.0.000636
- Lo S.W., Mellor K., Cohen R., Alonso A.R. Emergence of a multidrug-resistant and virulent Streptococcus pneumoniae lineage mediates serotype replacement after PCV13: an international whole-genome sequencing study. Lancet Microbe. 2022;3(10):e735–43. DOI: https://doi.org/10.1016/s2666-5247(22)00158-6
- Egorova E., Kumar N., Gladstone R.A., et al. Key features of pneumococcal isolates recovered in Central and Northwestern Russia in 2011–2018 determined through whole-genome sequencing. Microb. Genom. 2022;8(9):mgen000851. DOI: https://doi.org/10.1099/mgen.0.000851
- Beheshti M., Jabalameli F., Feizabadi M.M., et al. Molecular characterization, antibiotic resistance pattern and capsular types of invasive Streptococcus pneumoniae isolated from clinical samples in Tehran, Iran. BMC Microbiol. 2020;20(1):167. DOI: https://doi.org/10.1186/s12866-020-01855-y
- Zhou M., Wang L., Wang Z., et al. Molecular characterization of penicillin-binding protein2x, 2b and 1a of Streptococcus pneumoniae causing invasive pneumococcal diseases in China: a multicenter study. Front. Microbiol. 2022;13:838790. DOI: https://doi.org/10.3389/fmicb.2022.838790
- Ouldali N., Varon E., Levy C., et al. Invasive pneumococcal disease incidence in children and adults in France during the pneumococcal conjugate vaccine era: an interrupted time-series analysis of data from a 17-year national prospective surveillance study. Lancet Infect. Dis. 2021;21(1):137–47. DOI: https://doi.org/10.1016/S1473-3099(20)30165-1
- Andrejko K., Ratnasiri B., Lewnard J.A. Association of pneumococcal serotype with susceptibility to antimicrobial drugs: a systematic review and meta-analysis. Clin. Infect. Dis. 2022;75(1):131–40. DOI: https://doi.org/10.1093/cid/ciab852
- Balsells E., Dagan R., Yildirim I., et al. The relative invasive disease potential of Streptococcus pneumoniae among children after PCV introduction: a systematic review and meta-analysis. J. Infect. 2018;77(5):368–78. DOI: https://doi.org/10.1016/j.jinf.2018.06.004
- Amin-Chowdhury Z., Collins S., Sheppard C., et al. Characteristics of invasive pneumococcal disease caused by emerging serotypes after the introduction of the 13-valent pneumococcal conjugate vaccine in England: a prospective observational cohort study, 2014–2018. Clin. Infect. Dis. 2020;71(8):e235–43. DOI: https://doi.org/10.1093/cid/ciaa043
- Hansen C.B., Fuursted K., Valentiner-Branth P., et al. Molecular characterization and epidemiology of Streptococcus pneumoniae serotype 8 in Denmark. BMC Infect. Dis. 2021;21(1):421. DOI: https://doi.org/10.1186/s12879-021-06103-w
- Yamaguchi M. Investigation of pneumococcal virulence factors in the infection process. Nihon Saikingaku Zasshi. 2020;75(2): 173–83. DOI: https://doi.org/10.3412/jsb.75.173 (in Japanese)
Supplementary files