Current data on the circulation of the Q fever pathogen in the Republic of Guinea
- Authors: Naidenova E.V.1, Zakharov K.S.1, Agafonov D.A.1, Kartashov M.Y.2, Senichkina A.M.1, Khalilov E.S.3, Ibrahim A.B.4, Bah M.B.4, Nourdine I.4,5, Tokarevich N.K.3, Boumbaly S.4,5, Sidime Y.6, Shcherbakova S.A.1, Kutyrev V.V.1
-
Affiliations:
- Russian Research Anti-Plague Institute «Microbe»
- State Scientific Center of Virology and Biotechnology «Vector»
- Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology
- Research Institute of Applied Biology of Guinea
- Virology Research Center
- Institute of Veterinary Medicine
- Issue: Vol 101, No 5 (2024)
- Pages: 606-618
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18611
- DOI: https://doi.org/10.36233/0372-9311-557
- EDN: https://elibrary.ru/tezljr
- ID: 18611
Cite item
Abstract
Background. Q fever is the one of the best-studied zoonoses, which is widespread throughout almost the entire territory of Africa, excluding the territory of the Sahara. However, the current data on the incidence of coxiellosis and the circulation of Coxiella burnetii on this continent are limited and vary according different sources.
In 1980–1990, the Soviet-Guinean Research Virology and Microbiology Laboratory conducted studies to estimate the distribution of the Q fever pathogen, assess the herd immunity in humans and identify specific antibodies in the sera of livestock. However, in subsequent years, the research was suspended.
The aim of this study is to obtain up-to-date data on the distribution of C. burnetii in all landscape and geographical zones of the Republic of Guinea.
Materials and methods. The study was carried out in the laboratory of the Russian-Guinean Center for Epidemiology and Prevention of Infectious Diseases (Kindia, Republic of Guinea). The study involved 332 sera of febrile patients and 3156 sera from practically healthy volunteers, 1074 blood samples of livestock, 1648 suspensions of ticks, 319 specimens of small mammals and 298 of bats. The study was carried out using ELISA and PCR methods, selected samples were subjected to in-depth genetic analysis.
Results and discussion. The study of the distribution of C. burnetii on the territory of all landscape-geographical zones of the Republic of Guinea was carried out. For the first time, an officially registered case of human Q fever case has been identified. The role of livestock, small mammals and bats in the circulation of the pathogen has been established. It has been shown that the main vectors in Guinea are ixodid ticks of the Amblyomma variegatum, Hyalomma truncatum and Rhipicephalus decoloratus species. Employing molecular methods, C. burnetii strains carrying the QpH1 plasmid, capable of causing diseases in humans and animals were identified. For the first time, the complete nucleotide sequence of 16S rRNA of the Q fever pathogen (OQ152497–OQ152500) identified on the territory of Guinea was determined and registered in the GenBank database.
Conclusion. Taking into account the high epidemiological significance of Q fever, the study of the specifics of C. burnetii circulation in Guinea remains an urgent task. Regular monitoring and assessment of risk factors for diseases caused by coxiella are necessary for the development of an algorithm for laboratory diagnosis and recommendations for clinicians.
Full Text
Introduction
Q fever or coxiellosis is a common natural focal disease for humans and animals, the etiological agent of which is the bacteria Coxiella burnetii (family Legionellaceae, class Gammaproteobacteria), characterized by polymorphism of the clinical picture in humans and various mechanisms of transmission of the pathogen. In natural foci, the main vector of C. burnetii is considered to be ixodes, less frequently argas ticks, and the reservoir — wild mammals, among farm animals — small and large cattle [1, 2].
The disease in humans runs in the form of fever with general toxic symptoms with possible transition to a chronic form. Due to the widespread infection, a variety of transmission routes (contact, food, airborne and dust), Q fever is an important medical and social problem worldwide.
Furthermore, this infectious disease has an important veterinary significance, as it causes reproductive disorders (abortions and stillbirths) in small and large cattle, which causes significant economic losses, especially in those regions where livestock breeding is the main branch of agricultural production [2].
Coxiellosis is one of the best-studied zoonoses within Africa. There is emerging evidence that C. burnetii infection is responsible for non-malarial febrile illnesses and community-acquired pneumonia in many African countries. However, current information on the incidence of Q fever and circulation of C. burnetii in this continent is limited and heterogeneous [3].
Available publications on previous studies indicate a fairly high level of detection of specific antibodies to the pathogen in the population of some African countries [4–9]. For example, the study of blood serum samples from residents of some settlements of the Sin-Saloum region (Republic of Senegal) showed that specific antibodies to C. burnetii were detected in 3.7–24.8% of samples (depending on the place of residence of the subjects) [5]. Similar data were obtained during serosurvey in pastoralist communities in Marsabit County, northern Kena, when positive results were obtained in 13.2% of cases [6]. Males in Kenya were significantly more likely than females to have immunologic markers identified [7].
It has been shown that pastoralist tribes are at the highest risk of contracting coxiellosis because of their nomadic lifestyle and well-preserved traditions, which increase the likelihood of eating unboiled dairy products and raw meat from infected animals. It is also possible that infection occurs through contact with urine, feces, and blood of infected animals, as well as amniotic fluid after abortion or premature birth [6–9].
It is also known that Q fever causes significant losses among not only farm animals but also wild animals such as antelopes, giraffes, lions and cheetahs, causing irreparable damage to the numbers of these rare mammals [10].
This infectious disease is also becoming relevant as a “travelers' disease” due to the popularization of the African continent and rapidly developing tourism in this territory. An outbreak of Q fever was described in the literature, when during a safari in a natural park in Kenya, 4 (8%) out of 50 participants of the tourist route were infected, which caused importation of cases to Europe [11].
Most of the information published in the open press on the circulation of the Q fever pathogen relates to East Africa. There is much less information on the situation in the western part of the continent. There are data on studies conducted in natural foci of coxiellosis in Ghana, Nigeria, Mali [12–14], as well as in Senegal, where in 2023 two new genotypes of C. burnetii were identified in samples from small mammals, the pathogenicity of which remains to be studied [15].
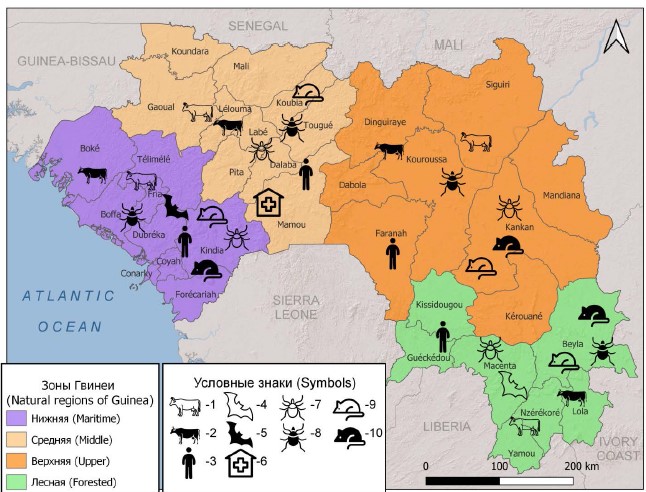
The Republic of Guinea is located in West Africa, on the coast of the Atlantic Ocean. The population of the country, as of the end of March 2024, is about 14.5 million people [16]. Based on geographical and natural-climatic features, the territory of the state is conditionally divided into 4 landscape-geographical zones: Lower (Maritime), Middle, Upper and Forested (Figure) [17].
Landscape and geographical zones of the Republic of Guinea, where markers of the Q fever pathogen were identified.
1 — DNA in blood sera of cattle; 2 — IgG antibodies in blood sera of cattle; 3 — IgG antibodies in blood sera of residents; 4 — DNA in suspensions of bat organs; 5 — antigens in suspensions of bat organs; 6 — cases of human disease; 7 — DNA in suspensions of ixodes ticks; 8 — antigens in suspensions of ixodes ticks; 9 — DNA in suspensions of organs of small mammals; 10 — antigens in suspensions of organs of small mammals.
In 1980–1990, the Soviet-Guinean Research Virology and Microbiology Laboratory conducted a large number of studies to investigate the spread of the Q fever pathogen in the Republic of Guinea, including data on the immunity of the country's population and the detection of specific antibodies in the blood sera of farm animals [18]. In the following decades, due to the prevailing economic and political conditions, research was suspended, and the significance of Q fever in the overall morbidity has not been determined. In 2017, as part of the research work of the Russian-Guinean Center for Epidemiology and Prevention of Infectious Diseases (hereinafter referred to as the Center), established on the basis of orders of the Government of the Russian Federation on the premises of the Institute of Applied Biology of Guinea (Kindia), research was continued [19–21].
The aim of the study was to obtain up-to-date data on the distribution of C. burnetii and the features of natural foci of Q fever in different landscape and geographical zones of Guinea.
Materials and methods
Collection of samples of clinical and biological material and subsequent diagnostic work was carried out on the basis of the Center's laboratory by Russian and Guinean specialists, guided by the requirements of sanitary rules and regulations 3.3686-21 “Sanitary and Epidemiological Requirements for the Prevention of Infectious Diseases”.
Human blood serum
Blood serum of practically healthy people was collected in regional hospitals in Guinea and by local specialists. Fasting blood sampling was performed in the morning hours from the ulnar vein in the amount of 5–10 ml into a disposable sterile vacuum tube with a clot activator in compliance with the rules of asepsis.
Then the obtained samples were delivered to the laboratory of the Center in compliance with the rules of biological safety and temperature regime. All serum samples were preliminarily analyzed by immunochromatographic analysis for detection of malaria plasmodium antigens with the SDBIOLINE Malaria Ag P.f./Pan reagent kit (Standart Diagnostics, Inc.). To exclude the possibility of nonspecific reactions, samples containing antigens of malaria pathogens were not included in subsequent studies.
A panel of 3156 sera from virtually healthy individuals living in all 4 landscape-geographic zones of Guinea was compiled to study the herd immunity to the Q fever pathogen in the Guinean population. Different age groups were included: 1519 (48.2%) women and 1637 (51.8%) men. The work was carried out using the enzyme-linked immunosorbent assay (ELISA) reagent kit “Enzyme-linked immunosorbent assay for detection of class G antibodies to C. burnetii antigens” (Pasteur Research Institute of Epidemiology and Microbiology, Russia). The sera were tested at a dilution of 1 : 100.
To clarify the possibility of human disease with Q fever, 332 sera from febrile patients who sought medical help in regional hospitals in Guinea with complaints of prolonged fever and other symptoms not excluding coxiellosis were collected and examined. The material was also collected by Guinean specialists using the methodology described above. For molecular genetic studies, blood was also collected in an amount of 5–10 ml in a disposable sterile vacuum tube with 3.8% sodium citrate.
Samples were examined by polymerase chain reaction (PCR) to detect C. burnetii DNA with the AmpliSens Coxiella burnetii-FL reagent kit (Central Research Institute of Epidemiology, Russia) and ELISA using the diagnostic kit Coxiella burnetii ELISA IgM (Vircell) to detect IgM antibodies to the pathogen.
The study of samples of clinical and biological material from humans was conducted with informed consent of patients, for minors — with the permission of parents (official representatives). The study protocol was approved by the decision of the National Ethical Committee of the Ministry of Health of the Republic of Guinea (protocol No. 129/CNERS/16 of August 31, 2015).
Blood serum of farm animals
Blood samples were obtained at slaughterhouses, using generally accepted methods, from adult animals (more than 1.5 years old) without signs of infectious diseases after their examination by a veterinarian. In total, a panel of 1074 cattle blood samples was formed for the work. The obtained sera were tested by ELISA and PCR methods using “ID Screen Q Fever Indirect Multi-species” reagent kits for detection of IgG antibodies specific to C. burnetii (ID Screen), which is recommended as a veterinary assay, and C. burnetii DNA was detected by the AmpliSens Coxiella burnetii-FL reagent kit (Central Research Institute of Epidemiology, Russia).
Tick suspensions
Ixodes ticks were collected in all 4 landscape-geographical zones of Guinea. Ectoparasites were removed manually, using personal protective equipment, from humans, farm animals, domestic and stray dogs and cats, small mammals, and reptiles. A total of 4709 specimens of ticks were collected during the studies, which were classified into 11 species based on morphological features: Amblyomma variegatum Fabricius, 1794; Haemaphysalis leachi Audouin, 1826; Hyalomma rufipes Koch, 1844; Hyalomma truncatum Koch, 1844; Rhipicephalus (Boophilus) decoloratus Koch, 1844; Rhipicephalus (Boophilus) geigyi Aeschliman & Morel, 1965; Rhipicephalus (Boophilus) annulatus Say, 1821; Rhipicephalus (Boophilus) microplus Canestrini, 1888; Rhipicephalus lunulatus Neumann, 1907; Rhipicephalus sanguineus Latreille, 1806; Rhipicephalus senegalensis Koch, 1844 according to the identification guide [22]. Further, 1648 pools were formed taking into account species, sex, developmental phase and fatness of individual mites, as well as places of ectoparasite collection. Ectoparasites were washed twice with 70% ethanol to remove external contaminants and external microflora. Samples were prepared using a TissueLyser II laboratory homogenizer (Qiagen) in 500 µl of sterile phosphate-salt buffer solution.
The obtained material was tested by PCR and ELISA methods using the AmpliSens Coxiella burnetii-FL reagent kit (Central Research Institute of Epidemiology) and ELISA-Qu-antigen (kit N1) test system for detection of Coxiella burnetii antigens (Pasteur Research Institute of Epidemiology and Microbiology), respectively. Some samples containing both DNA and antigens of C. burnetii were analyzed using high-throughput sequencing on the Ion S5 platform (Thermo Scientific); further reads were mapped to the 16S rRNA sequence from the NCBI GenBank database using the BWA algorithm [23]. The plasmid profile with specific primers to the QpH1, QpRS and QpDV plasmid loci was also studied.
Small mammal organ suspensions
During epizootological monitoring on the territory of Guinea, 319 specimens of small mammals (Rodentia, Eulipotyphla) were captured. The species spectrum of rodents was represented by 13 species: Arvicanthis ansorgei Thomas, 1910 (Sudan grass mouse); Heliosciurus gambianus Ogibly, 1835 (Gambian squirrel); Cricetomys gambianus Waterhouse, 1840 (Gambian hamster rat); Lemniscomys striatus Linnaeus, 1758 (striped mouse); Lophuromys sikapusi Temminck, 1853 (rusty-bellied stiff-necked mouse); Mastomys erythroleucus Temminck, 1853 (Guinean multispotted mouse); Mastomys natalensis A. Smith, 1834 (Natal mouse); Mus minutoides A. Smith, 1834 (dwarf mouse); Mus musculoides Temminck, 1853 (Temminck's mouse); Praomys daltoni Thomas, 1892 (Dalton's mouse); Rattus rattus Linnaeus, 1758 (black rat); Crocidura olivieri Lesson, 1827 (African giant whitetooth); Crocidura sp. Wagler, 1832 (whitetooths) [24–28].
We obtained 298 specimens of bats (Chiroptera) of 14 species: Eidolon helvum Kerr, 1792 (palmate wing); Epomophorus gambianus Ogilby, 1835 (great epaulet wing); Lissonycteris angolensis Bocage, 1898 (Angolan flying dog); Rousettus aegyptiacus E. Geoffroy, 1810 (Egyptian flying dog); Chaerephon pumillus Cretzschmar, 1830 (dwarf folding spoonbill); Mops condylurus A. Smith, 1833 (Angolan folding spoonbill); Hipposideros caffer Sundevall, 1846 (South African leafhopper); Hipposideros jonesi Hayman, 1947 (Jones' leafhopper); Hipposideros ruber Noack, 1893 (red leafhopper); Nycteris hispida Schreber, 1775 (shaggy slitmouth); Neoromicia guineensis Bocage, 1889 (Guinean leatherback); Scotophilus dinganii A. Smith, 1833 (African smooth-nosed platypus); Scotophilus leucogaster Cretzschmar, 1830 (white-bellied house smooth-nosed platypus); Rhinolophus alcyone Temminck, 1853 (Ghanaian horseshoe) [24-27].
Mammals were dissected in compliance with the rules of biological safety no later than 3 h after capture. In the absence of such an opportunity, animal carcasses were frozen at –20ºС and delivered to the laboratory, where they were subsequently dissected and organ samples were taken. Combined suspensions of lungs and kidneys were used as material for work, in which markers of the coxiellosis pathogen (DNA and antigens) were searched with the reagent kits mentioned above.
The authors confirm compliance with institutional and national standards for the use of laboratory animals in accordance with the “Consensus author guidelines for animal use” (IAVES July 23, 2010). The study was approved by the Bioethics Commission of the Russian Anti-Plague Institute “Microbe” (protocol No. 8 of November 21, 2023).
During statistical processing of the material we calculated the proportion of detected pathogen markers in each sample, 95% confidence intervals (CI) for the proportions using Wilson's method.
Results and discussion
Identification of human cases of Q fever in Guinea
To date, there has been no detection of suspected cases of Q fever in Guinea. This can be explained by the lack of alertness of health workers in local hospitals to this infectious disease. In this regard, a case of the disease reported for the first time in the country is of great interest.
Patient D., 28 years old, living in Mamou (Middle Guinea), was admitted to the infectious disease department of a regional hospital. At hospitalization, the patient had prolonged (more than 1 month), subfebrile fever, muscle and joint pains and breathing difficulties. The results of immunochemistry and microscopy tests for detection of malaria pathogens were negative.
For further differential diagnosis, the patient's blood serum was sent to the Center's laboratory. The obtained material was analyzed by reverse transcription PCR to detect RNA of Ebola, yellow fever, West Nile, dengue, Zika, Crimean-Congo hemorrhagic fever, hepatitis C, 16S RNA of leptospirosis pathogens, DNA of hepatitis B virus, pathogens of Q fever and rickettsioses. Based on the results, C. burnetii DNA was detected in the patient's serum. The material was also tested using ELISA and specific IgM antibodies to coxiellae were detected in a titer of 1 : 400. Subsequently, the obtained blood sample was used to determine the nucleotide sequence of the DNA of the Q fever pathogen. Partial sequencing with specific primers revealed 99% identity of the tested sample with the genome of C. burnetii. Phylogenetic analysis using the BLAST algorithm1 showed 96% homology with strains isolated in Namibia.
When interviewed, patient D. mentioned close contact with farm animals, specifically owned cattle, and also indicated that there had been cases of spontaneous abortions of cattle in the patient's village.
Neither genetic markers of C. burnetii nor specific IgM antibodies to coxiellae were detected in the remaining serum samples from febrile patients transported from Guinea hospitals.
Determination of the herd immunity of Guineans to the Q fever pathogen
In the present study, specific IgG antibodies to the Q fever pathogen were detected in sera of participants of all age groups living in different zones of Guinea, and there was no dependence of the antibody detection rates on the sex of the examined persons (Table 1). In the whole country, specific immunoglobulins to C. burnetii were registered in 366 out of 3156 serum samples, which amounted to 11.6% (95% CI 10.5-12.8).
Table 1. Identification of specific IgG antibodies to the Q fever pathogen in the blood sera of residents of the Republic of Guinea
Age, years | Number of samples | ||||||
total | men | women | |||||
total | positive | % (95% CI) | total | positive | % (95% CI) | ||
< 10 | 197 | 108 | 8 | 7.4 (3.8–13.9) | 89 | 6 | 6.7 (3.1–13.9) |
10–20 | 498 | 275 | 26 | 9.4 (6.5–13.4) | 223 | 27 | 12.1 (8.4–17.0) |
20–30 | 699 | 340 | 32 | 9.4 (6.7–12.9) | 359 | 29 | 8.1 (5.7–11.4) |
30–40 | 595 | 300 | 39 | 13.0 (9.7–17.3) | 295 | 33 | 11.2 (8.1–15.3) |
40–50 | 559 | 285 | 35 | 12.3 (8.9–16.6) | 277 | 29 | 10.5 (7.4–14.6) |
50–60 | 339 | 230 | 37 | 16.1 (11.9–21.4) | 109 | 26 | 23.8 (16.8–32.6) |
> 70 | 269 | 102 | 20 | 19.6 (13.1–28.3) | 167 | 19 | 11.4 (7.4–17.1) |
Total | 3156 | 1637 | 197 | 12.1 (10.5–13.7) | 1519 | 169 | 11.3 (9.6–12.8) |
Determination of the herd immunity of farm animals to the Q fever pathogen
One of the indicators of circulation of pathogens of naturally occurring infectious diseases in a certain area is the detection of specific IgG immunoglobulins in blood sera of farm animals living in this region.
As a result of this work, antibodies to the Q fever pathogen were detected in 172 samples, which amounted to 16.0% (95% CI 13.9-18.3). Positive results were recorded in all landscape-geographical zones).
Detection of specific markers (DNA and antigens) of the Q fever pathogen in tick suspensions
PCR and ELISA methods were used to examine suspensions of ixodid ticks of various species, which are the main vectors of C. burnetii, collected in all landscape and geographical zones of Guinea. The pathogen DNA was detected in 294 (17.9%) samples and antigen in 307 (18.7%). Positive findings were noted among all ixodid tick species represented in the work, but the majority were attributed to Am. variegatum, Hy. truncatum, Rh. decoloratus (Table 2).
Table 2. Identification of C. burnetii markers in suspensions of ixodid ticks of different species collected in the territory of the Republic of Guinea
Tick species | Number of samples (copies) | The number of positive samples; % (95% СI) | |
PCR | ELISA | ||
Am. variegatum | 872 (2493) | 159; 18.2 (15.8–20.9) | 193; 22.1 (19.5–25.0) |
Ha. leachi | 16 (56) | 0 | 0 |
Hy. truncatum | 52 (95) | 14; 26.9 (16.8–40.3) | 16; 30.8 (19.9–44.3) |
Rh. annulatus | 58 (161) | 36; 62.1 (49.2–73.4) | 29; 50 (37.5–62.5) |
Rh. decoloratus | 391 (1104) | 47; 12 (9.2–15.6) | 37; 9.5 (6.9–12.8) |
Rh. geigyi | 210 (668) | 36; 17.1 (12.7–22.8) | 29; 13.8 (9.8–19.1) |
Rh. microplus | 18 (47) | 0 | 1; 5.6 (1.0–25.8) |
Rh. sanguineus | 10 (42) | 0 | 1; 10 (1.8–40.4) |
Rh. senegalensis | 18 (29) | 2; 11.1 (3.1–32.8) | 1; 5.6 (1.0–25.8) |
Rh. lunulatus | 2 (11) | 0 | 0 |
Hy. rufipes | 1 (3) | 0 | 0 |
Total | 1648 (4709) | 294; 17.8 (16.1–19.8) | 307; 18.6 (16.8–20.6) |
Analysis of plasmid profiles is an important tool for studying the spread of Q fever and determining the type of the pathogen. For genetic typing of C. burnetii, a panel of 20 samples from different tick species was created, in which both DNA (Ct level not more than 15) and antigen of the pathogen were detected simultaneously. PCR with specific primers to the QpH1, QpRS and QpDV plasmid loci revealed the presence of QpH1 alone in 5 samples. Sequences of QpRS and QpDV plasmid fragments were not detected in any of the samples. In the course of analyzing the results obtained and literature data, it was found that strains carrying the QpH1 plasmid are widespread in Equatorial Africa and are capable of causing diseases in humans and animals [29, 30], which does not exclude the possibility of their circulation in Guinea.
The generated panel of samples was analyzed using high-throughput sequencing methods on the Ion S5 platform (Thermo Scientific). As a result, the nucleotide sequence of 16S rRNA of the Q fever pathogen was determined in 8 samples, which coincides 99.9% with the reference strain presented in the NCBI GenBank database. When comparing the obtained consensus sequence with the NCBI BLAST database, C. burnetii strains isolated in Namibia were found to show homology (96%) with the studied sample. Furthermore, the obtained reads were classified by the kraken2 algorithm using the Greengenes 16S RNA base, which also showed that the investigated sample belonged to the C. burnetii species. Some of the obtained 16S rRNA nucleotide sequences with the highest quality reads were deposited in the international GenBank database under the numbers OQ152497-OQ152500.
Detection of specific markers (DNA and antigens) of the Q fever pathogen in organ suspensions of small mammals
As a result of PCR and ELISA tests, markers of the Q fever pathogen were detected in the material collected in all zones of Guinea. Antigen was detected in 0.9% of samples and DNA in 5.1%. The maximum number of positive findings was obtained when samples from Mastomys erythroleucus rodents was examined (Table 3). These data may indicate the participation of animals of this systematic group in the spread of C. burnetii in Guinea.
Table 3. Identification of C. burnetii markers in suspensions of organs of small mammals of different species collected on the territory of the Republic of Guinea
Small mammal species | Number of samples (copies) | The number of positive samples; % (95% СI) | |
PCR | ELISA | ||
Arvicanthis ansorgei | 3 | 0 | 0 |
Heliosciurus gambianus | 4 | 0 | 0 |
Cricetomys gambianus | 7 | 1; 14.3 (2.6–51.3) | 0 |
Lemniscomys striatus | 3 | 0 | 0 |
Lophuromys sikapusi | 1 | 0 | 0 |
Mastomys erythroleucus | 124 | 8; 6.5 (3.3–12.2) | 2; 1.6 (0.4–5.7) |
Mastomys natalensis | 32 | 2; 6.3 (1.7–20.1) | 0 |
Mus minutoides | 5 | 0 | 0 |
Mus musculoides | 16 | 1; 6.3 (1.1–28.3) | 0 |
Praomys daltoni | 6 | 0 | 0 |
Rattus rattus | 96 | 4; 4.2 (1.6–10.2) | 1; 1.0 (0.2–5.7) |
Crocidura olivieri | 7 | 0 | 0 |
Crocidura sp. | 15 | 0 | 0 |
Total | 319 | 16; 5.0 (3.1–8.0) | 3; 0.9 (0.3–2.7) |
Detection of specific markers (DNA and antigens) of the Q fever pathogen in organ suspensions of bats
Examination of pooled samples of lungs and kidneys obtained from bats revealed both C. burnetii antigens (1% of cases) and DNA (2%). The majority of positive samples were formed from organs of Scotophilus leucogaster (Table 4). Representatives of this species are widely distributed throughout sub-Saharan Africa. The data obtained confirm the role of bats in the circulation of the Q fever pathogen, which indicates the need for additional studies to determine the role of these mammals in the ecology of the pathogen.
Table 4. Identification of C. burnetii markers in organ suspensions of bats of various species collected on the territory of the Republic of Guinea
Bat species | Number of samples (copies) | The number of positive samples; % (95% СI) | |
PCR | ELISA | ||
Eidolon helvum | 1 | 0 | 0 |
Epomophorus gambianus | 4 | 0 | 0 |
Lissonycteris angolensis | 4 | 0 | 0 |
Rousettus aegyptiacus | 15 | 0 | 0 |
Chaerephon pumillus | 3 | 0 | 0 |
Mops condylurus | 26 | 1; 3.8 (0.7–18.9) | 0 |
Hipposideros caffer | 32 | 0 | 0 |
Hipposideros jonesi | 25 | 0 | 0 |
Hipposideros ruber | 41 | 1; 2.4 (0.4–12.6) | 1; 2.4 (0.4–12.6) |
Nycteris hispida | 5 | 0 | 0 |
Neoromicia guineensis | 23 | 1; 4.3 (0.8–21.0) | 0 |
Scotophilus dinganii | 1 | 0 | 0 |
Scotophilus leucogaster | 117 | 3; 2.6 (0.9–7.3) | 2; 1.7 (0.5–6.0) |
Rhinolophus alcyone | 1 | 0 | 0 |
Total | 298 | 6; 2.0 (0.9–4.3) | 3; 1.0 (0.3–2.9) |
Conclusion
As a result of this work, the distribution of C. burnetii in all landscape-geographical zones of Guinea was studied, including the first case of human case of Q fever identified and laboratory confirmed (Table 5). The role of farm animals, small mammals and bats in the circulation of C. burnetii has been established. It was shown that the main vectors of the pathogen on the territory of Guinea are ixodes ticks of Am. variegatum, Hy. truncatum and Rh. decoloratus species. Molecular genetic studies of the material collected in Guinea revealed C. burnetii strains carrying the QpH1 plasmid, which are capable of causing disease in humans and animals, and for the first time for this region the complete nucleotide sequence of the 16S rRNA gene of the Q fever pathogen was determined and registered in the international GenBank database.
Table 5. Identification of markers of the Q fever pathogen in various materials collected on the territory of the Republic of Guinea
Type of the studied samples | Number of samples | The number of positive; % (95% CI) | ||
PCR | ELISA | |||
DNA | antigen | IgG | ||
Lower (Maritime) Guinea | ||||
Blood serum of practically healthy people | 943 | N. i. | N. i. | 106; 11.2 (9.4–13.4) |
Blood serum of cattle | 371 | 6; 1.6 (0.7–3.5) | N. i. | 64; 17.2 (13.7–21.4) |
Suspensions of ticks | 624 | 111; 17.8 (15.0–21.0) | 131; 21.0 (18.0–24.4) | N. i. |
Suspensions of organs of small mammals | 149 | 8; 5.4 (2.7–10.2) | 0; 0 (0–2.5) | N. i. |
Suspensions of bat organs | 107 | 0; 0 (0–3.4) | 3; 2.8 (0.9–7.9) | N. i. |
Middle Guinea | ||||
Blood serum of practically healthy people | 778 | N. i. | N. i. | 82; 10.5 (8.6–12.8) |
Blood serum of cattle | 257 | 3; 1.2 (0.4–3.4) | N. i. | 49; 19.2 (14.7–24.3) |
Suspensions of ticks | 402 | 71; 17.7 (14.2–21.7) | 77; 19.1 (15.6–23.3) | N. i. |
Suspensions of organs of small mammals | 55 | 3; 5.4 (1.9–14.8) | 0; 0 (0–6.5) | N. i. |
Suspensions of bat organs | 61 | 0; 0 (0–5.8) | 0; 0 (0–5.8) | N. i. |
Upper Guinea | ||||
Blood serum of practically healthy people | 655 | N. i. | N. i. | 77; 11.8 (9.5–14.4) |
Blood serum of cattle | 182 | 2; 1.1 (0.3–3.9) | N. i. | 35; 13.2 (9.6–17.8) |
Suspensions of ticks | 245 | 53; 21.6 (16.9–27.2) | 28; 0.8 (0.3–2.9) | N. i. |
Suspensions of organs of small mammals | 43 | 2; 4.6 (1.3–15.4) | 1; 2.3 (0.4–12.1) | N. i. |
Suspensions of bat organs | 54 | 0; 0 (0–6.6) | 0; 0 (0–6.6) | N. i. |
Forest Guinea | ||||
Blood serum of practically healthy people | 780 | N. i. | N. i. | 101; 12.9 (10.7–15.5) |
Blood serum of cattle | 264 | 3; 1.1 (0.4–3.3) | N. i. | 35; 13.2 (9.6–17.8) |
Suspensions of ticks | 377 | 59; 15.6 (12.3–19.7) | 61; 16.2 (12.8–20.2) | N. i. |
Suspensions of organs of small mammals | 72 | 3; 4.2 (1.4–11.5) | 2; 2.8 (0.8–9.5) | N. i. |
Suspensions of bat organs | 76 | 6; 7.9 (3.7–16.1) | 0; 0 (0–4.8) | N. i. |
General by country | ||||
Blood serum of practically healthy people | 3156 | N. i. | N. i. | 366; 11.6 (10.5–12.7) |
Blood serum of cattle | 1074 | 14; 1.3 (0.8–2.1) | N. i. | 172; 16.0 (13.9–18.3) |
Suspensions of ticks | 1648 | 294; 17.8 (16.1–19.8) | 307; 18.6 (16.8–20.6) | N. i. |
Suspensions of organs of small mammals | 319 | 16; 5.0 (3.1–8.0) | 3; 0.9 (0.3–2.7) | N. i. |
Suspensions of bat organs | 298 | 6; 2.0 (0.9–4.3) | 3; 1.0 (0.3–2.9) | N. i. |
Note. N. i. — not investigated.
Our study allowed us to expand the data on the circulation and distribution of C. burnetii in West Africa. The study of the specifics of C. burnetii circulation in Guinea remains an urgent task. Systematically obtained data on the detection of the pathogen and assessment of risk factors for outbreaks of diseases caused by coxiellae are necessary for the development of an algorithm for laboratory diagnosis and recommendations for clinicians. Regular monitoring of the spread of Q fever, carried out with the participation of both medical and veterinary services in Guinea, will make it possible to forecast the epidemiological situation and coordinate preventive (anti-epidemic) measures within the framework of the One Health concept2.
1 URL: https://blast.ncbi.nlm.nih.gov/Blast.cgi
2 WHO. One health. URL: https://www.who.int/health-topics/one-health#tab=tab_1 (дата обращения: 26.05.2024).
About the authors
Ekaterina V. Naidenova
Russian Research Anti-Plague Institute «Microbe»
Author for correspondence.
Email: katim2003@mail.ru
ORCID iD: 0000-0001-6474-3696
senior researcher, Department of infectious disease diagnostics
Russian Federation, SaratovKirill S. Zakharov
Russian Research Anti-Plague Institute «Microbe»
Email: katim2003@mail.ru
ORCID iD: 0000-0002-4726-309X
senior researcher, Department of infectious disease epidemiology
Russian Federation, SaratovDmitry A. Agafonov
Russian Research Anti-Plague Institute «Microbe»
Email: katim2003@mail.ru
ORCID iD: 0000-0001-9273-6063
старший научный сотрудник отдела диагностики инфекционных болезней ФКУН Российский противочумный институт «Микроб» Роспотребнадзора, к.б.н.
Russian Federation, SaratovMikhail Yu. Kartashov
State Scientific Center of Virology and Biotechnology «Vector»
Email: katim2003@mail.ru
ORCID iD: 0000-0002-7857-6822
Cand. Sci. (Biol.), senior researcher, Department of molecular virology of flaviviruses and viral hepatitis
Russian Federation, Kol’tsovoAislu M. Senichkina
Russian Research Anti-Plague Institute «Microbe»
Email: katim2003@mail.ru
ORCID iD: 0000-0003-1026-2680
Cand. Sci. (Biol.), senior researcher, Department of diagnostics of infectious diseases
Russian Federation, SaratovErik S. Khalilov
Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology
Email: katim2003@mail.ru
ORCID iD: 0000-0002-0599-4302
junior researcher, Laboratory of zoonotic infections
Russian Federation, St. PetersburgAbdoul B. Ibrahim
Research Institute of Applied Biology of Guinea
Email: katim2003@mail.ru
ORCID iD: 0009-0003-2547-8705
researcher, Laboratory of viral hemorrhagic fevers of Guinea
Guinea, KindiaMamadou B. Bah
Research Institute of Applied Biology of Guinea
Email: katim2003@mail.ru
ORCID iD: 0000-0002-4565-269X
researcher
Guinea, KindiaIbrahim Nourdine
Research Institute of Applied Biology of Guinea; Virology Research Center
Email: katim2003@mail.ru
ORCID iD: 0000-0002-2970-9676
Cand. Sci. (Biol.), researcher, Laboratory of viral hemorrhagic fevers of Guinea
Guinea, Kindia; ConakryNikolai K. Tokarevich
Saint Petersburg Pasteur Research Institute of Epidemiology and Microbiology
Email: katim2003@mail.ru
ORCID iD: 0000-0001-6433-3486
D. Sci. (Med.), Professor, Head, Laboratory of zooanthroponotic infections
Russian Federation, St. PetersburgSanaba Boumbaly
Research Institute of Applied Biology of Guinea; Virology Research Center
Email: katim2003@mail.ru
ORCID iD: 0000-0002-4506-6033
Cand. Sci. (Biol.), Professor, Director, Laboratory of viral hemorrhagic fevers of Guinea
Guinea, Kindia; ConakryYusuf Sidime
Institute of Veterinary Medicine
Email: katim2003@mail.ru
ORCID iD: 0000-0002-0742-0468
Cand. Sci. (Vet.), Professor, Director, Institute of Veterinary Medicine
Guinea, DalabaSvetlana A. Shcherbakova
Russian Research Anti-Plague Institute «Microbe»
Email: katim2003@mail.ru
ORCID iD: 0000-0003-1143-4069
D. Sci. (Biol.), Deputy director
Russian Federation, SaratovVladimir V. Kutyrev
Russian Research Anti-Plague Institute «Microbe»
Email: katim2003@mail.ru
ORCID iD: 0000-0003-3788-3452
D. Sci. (Med.), Professor, Academician of the RAS, Director, Russian Anti-Plague Institute "Microbe"
Russian Federation, SaratovReferences
- Онищенко Г.Г., Кутырева В.В., ред. Специфическая индикация патогенных биологических агентов. М.; 2014. Onishchenko G.G., Kutyrev V.V., eds. Specific Indication of Pathogenic Biological Agents. Moscow; 2014. EDN: https://elibrary.ru/qlnqhv
- Лукин Е.П., Мищенко О.А., Борисевич С.В. Лихорадка Ку в XXI в.: материал для подготовки лекции. Инфекционные болезни: новости, мнения, обучение. 2019;8(4):62–77. Lukin E.P., Mishchenko O.A., Borisevich S.V. Ku fever in the XXI century: material for preparing a lecture. Infectious Diseases: News, Opinions, Training. 2019;8(4):62–77. DOI: https://doi.org/10.24411/2305-3496-2019-14009, EDN: https://elibrary.ru/houbuj
- Vanderburg S., Rubach M.P., Halliday J.E., et al. Epidemiology of Coxiella burnetii infection in Africa: a OneHealth systematic review. PLoS Negl. Trop. Dis. 2014;8(4):e2787. DOI: https://doi.org/10.1371/journal.pntd.0002787
- Dupont H.T., Brouqui P., Faugere B., Raoult D. Prevalence of antibodies to Coxiella burnetti, Rickettsia conorii, and Rickettsia typhi in seven African countries. Clin. Infect. Dis. 1995;21(5):1126–33. DOI: https://doi.org/10.1093/clinids/21.5.1126
- Mediannikov O., Fenollar F., Socolovschi C., et al. Coxiella burnetii in humans and ticks in rural Senegal. PLoS Negl. Trop. Dis. 2010;4(4):e654. DOI: https://doi.org/10.1371/journal.pntd.0000654
- Muema J., Nyamai M., Wheelhouse N., et al. Endemicity of Coxiella burnetii infection among people and their livestock in pastoral communities in northern Kenya. Heliyon. 2022;8(10):e11133. DOI: https://doi.org/10.1016/j.heliyon.2022.e11133
- Mwololo D., Nthiwa D., Kitala P., et al. Sero-epidemiological survey of Coxiella burnetii in livestock and humans in Tana River and Garissa counties in Kenya. PLoS Negl. Trop. Dis. 2022;16(3):e0010214. DOI: https://doi.org/10.1371/journal.pntd.0010214
- Crump J.A., Morrissey A.B., Nicholson W.L., et al. Etiology of severe non-malaria febrile illness in Northern Tanzania: a prospective cohort study. PLoS Negl. Trop. Dis. 2013;7(7):e2324. DOI: https://doi.org/10.1371/journal.pntd.0002324
- Larson P.S., Espira L., Grabow C., et al. The sero-epidemiology of Coxiella burnetii (Q fever) across livestock species and herding contexts in Laikipia County, Kenya. Zoonoses Public Health. 2019;66(3):316–24. DOI: https://doi.org/10.1111/zph.12567
- Mangena M.L., Gcebe N., Thompson P.N., Adesiyun A.A. Q fever and toxoplasmosis in South African livestock and wildlife: a retrospective study on seropositivity, sporadic abortion, and stillbirth cases in livestock caused by Coxiella burnetii. BMC Vet. Res. 2023;19(1):168. DOI: https://doi.org/10.1186/s12917-023-03645-w
- Njeru J., Henning K., Pletz M.W., et al. Febrile patients admitted to remote hospitals in Northeastern Kenya: seroprevalence, risk factors and a clinical prediction tool for Q-Fever. BMC Infect. Dis. 2016;16:244. DOI: https://doi.org/10.1186/s12879-016-1569-0
- Dione M.M., Séry A., Sidibé C.A.K., et al. Exposure to multiple pathogens — serological evidence for Rift Valley fever virus, Coxiella burnetii, Bluetongue virus and Brucella spp. in cattle, sheep and goat in Mali. PLoS Negl. Trop. Dis. 2022;16(4):e0010342. DOI: https://doi.org/10.1371/journal.pntd.0010342
- Addo S.O., Bentil R.E., Baako B.O.A., et al. Occurrence of Rickettsia spp. and Coxiella burnetii in ixodid ticks in Kassena-Nankana, Ghana. Exp. Appl. Acarol. 2023;90(1-2):137–53. DOI: https://doi.org/10.1007/s10493-023-00808-0
- Kamani J., Baneth G., Gutiérrez R., et al. Coxiella burnetii and Rickettsia conorii: Two zoonotic pathogens in peridomestic rodents and their ectoparasites in Nigeria. Ticks Tick Borne Dis. 2018;9(1):86–92. DOI: https://doi.org/10.1016/j.ttbdis.2017.10.004
- Mangombi-Pambou J., Granjon L., Labarrere C., et al. New genotype of Coxiella burnetii causing epizootic q fever outbreak in rodents, Northern Senegal. Emerg. Infect. Dis. 2023;29(5):1078–81. DOI: https://doi.org/10.3201/eid2905.221034
- Черч Гаррисон Р.Дж. Западная Африка. Природная среда и ее хозяйственное использование. М.;1959. Church Harrison R.J. West Africa. A study of the environment and of man's use of it. New York;1957.
- Каливоги С., Буаро М.Е., Константинов О.К., Плотникова Л.Ф. Иммунная структура населения и домашних животных Гвинейской Республики в отношении риккетсиозов группы клещевой пятнистой лихорадки и лихорадки Ку. Медицинская паразитология и паразитарные болезни. 2013;(1):28–30. Kalivogi S., Boiro M.E., Konstantinov O.K., Plotnikova L.F. The immune structure of the population and domestic animals of the Republic of Guinea in relation to rickettsioses of the tick-borne spotted fever and Ku fever group. Medical Parasitology and Parasitic Diseases. 2013;(1):28–30. EDN: https://elibrary.ru/tvzcbv
- Найденова Е.В., Каливоги С., Карташов М.Ю. и др. Новые данные об уровне иммунной прослойки населения Гвинейской Республики к возбудителю лихорадки Ку. Инфекция и иммунитет. 2021;11(1):165–70. Naidenova E.V., Kalivogui S., Kartashov M.Y., et al. New data on the level of immune stratum against Q fever agent in population of the of Republic of Guinea. Russian Journal of Infection and Immunity. 2021;11(1):165–70. DOI: https://doi.org/10.15789/2220-7619-NDO-1485 EDN: https://elibrary.ru/wzsxmv
- Найденова Е.В., Захаров К.С., Карташов М.Ю. и др. Выявление генетических маркеров возбудителей природно-очаговых инфекционных болезней в пробах иксодовых клещей, собранных на территории Гвинейской Республики. Проблемы особо опасных инфекций. 2023;(4):115–24. Naidenova E.V., Zakharov K.S., Kartashov M.Yu., et al. Genetic Marker Detection of Natural-Focal Infectious Disease Pathogens in Samples of Ixodidae Ticks, Collected on the Territory of the Republic of Guinea. Problems of Particularly Dangerous Infections. 2023;(4):115–24. DOI: https://doi.org/10.21055/0370-1069-2023-4-115-124 EDN: https://elibrary.ru/lnomas
- Найденова Е.В., Карташов М.Ю., Шевцова А.П. и др. Определение уровня иммунной прослойки сельскохозяйственных животных к возбудителям зоонозных инфекционных болезней в Гвинейской Республике. Проблемы особо опасных инфекций. 2022;(2):101–6. Naidenova E.V., Kartashov M.Yu., Shevtsova A.P., et al. Identification of the farm animals immune to pathogens of zoonotic infectious diseases in the Republic of Guinea. Problems of Particularly Dangerous Infections. 2022;(2):101–6. DOI: https://doi.org/10.21055/0370-1069-2022-2-101-106 EDN: https://elibrary.ru/bssaaq
- Walker A.R., Bouattour A., Camicas J.L., et al. Ticks of Domestic Animals in Africa: A Guide to Identification of Species. Edinburgh;2014.
- Li H., Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009; 25(14):1754–60. DOI: https://doi.org/10.1093/bioinformatics/btp324
- Wilson D.E., Reeder D.M., eds. Mammal Species of the World. A Taxonomic and Geographic Reference. Baltimore;2005.
- Happold D.C.D., ed. Mammals of Africa. Volume III: Rodents, Hares and Rabbits. London;2013.
- Happold M., Happold D.C.D., eds. Mammals of Africa. Volume IV: Hedgehogs, Shrews and Bats. London;2013.
- Соколов В.Е. Пятиязычный словарь названий животных. Млекопитающие. Латинский — русский — английский — немецкий — французский. М.;1984. Sokolov V.E. A Five-Language Dictionary of Animal Names. Mammals. Latin — Russian — English — German — French. Moscow;1984.
- Luo S., Lu S., Fan H., et al. The Coxiella burnetii QpH1 plasmid is a virulence factor for colonizing bone marrow-derived murine macrophages. J. Bacteriol. 2021;203(9):e00588–20. DOI: https://doi.org/10.1128/jb.00588-20
- Панферова Ю.А., Фрейлихман О.А., Токаревич Н.К. и др. Детекция Coxiella burnetii в клещах, собранных с крупного рогатого скота, на территории некоторых провинций Гвинейской Республики. Эпидемиология и инфекционные болезни. 2019;24(5-6):234–9. Panferova Yu.A., Freylikhman О.А., Tokarevich N.K., et al. Detection of Coxiella burnetii in ticks collected from cattle in several provinces of the Republic of Guinea. Epidemiology and infectious diseases. 2019;24(5-6): 234–9. EDN: https://elibrary.ru/twsemw
Supplementary files