Analysis of production levels of InlA and InlB invasion factors in Listeria monocytogenes isolates collected in the Russian Federation
- Authors: Kalinin E.V.1, Chalenko Y.M.1, Safarova P.V.1, Fedorova V.A.1, Ermolaeva S.A.1
-
Affiliations:
- Gamaleya Research Center of Epidemiology and Microbiology
- Issue: Vol 100, No 5 (2023)
- Pages: 276-286
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18474
- DOI: https://doi.org/10.36233/0372-9311-397
- EDN: https://elibrary.ru/ucqwtu
- ID: 18474
Cite item
Abstract
Background. Listeria monocytogenes is characterized by the presence of epidemic hypervirulent clones. A key feature of L. monocytogenes is its capacity to invade non-professional phagocytic cells. Hypervirulent clones are strongly associated with the increased production and/or the presence of certain isoforms of invasion factors InlA and InlB.
The purpose of the study is to create a test system for InlA and InlB detection and to measure the InlA and InlB production levels in L. monocytogenes isolates belonging to clonal groups with different virulence potential.
Materials and methods. The study was performed using 32 L. monocytogenes strains belonging to epidemic clones ECII, ECIV, ECVII (clonal complexes CC1, CC2, CC7) and hypovirulent clonal complex CC9. Sequencing of inlA and inlB genes was performed. The indirect enzyme-linked immunosorbent assay was used to analyze the production levels of InlA and InlB proteins.
Results. The variability of InlA was revealed among strains belonging to the same clonal complex: 3 InlA isoforms were identified among strains belonging to CC7; out of 8 strains belonging to CC9, one strain had a stop codon in the inlA gene, leading to the loss of function of the InlA protein. The differences between inlB alleles correlated with the specificity of strains belonging to a certain clonal complex. Differences in production levels of invasion factors were measured. In strains belonging to CC9, the InlA production level was 2.5 times as low compared to strains belonging to CC1, CC2, and CC7. In strains belonging to phylogenetically related CC1 and CC2, the InlB production level was on average 4 times as high compared to strains belonging to CC7 and CC9.
Conclusion. The obtained results confirm the variability of major invasion factors both among clonal complexes and strains of the same complex. The increased production of invasion factors InlA and InlB correlates with the potential virulence of strains.
Full Text
Introduction
Listeria monocytogenes causes a dangerous foodborne infection with a high (around 30%) mortality rate in high-risk individuals as well as in domestic and wild animals [1, 2]. The genetic structure of the L. monocytogenes species is divided into 4 phylogenetic lineages [3].
Phylogenetic lineage I includes clonal complexes (CCs) CC1, CC2, CC4, and CC6 most frequently associated with listeriosis in humans [4]. Using different methods, CC1 and CC2 strains were assigned to epidemic clones ECII and ECIV, respectively, which were involved in large listeriosis outbreaks in different countries, including Russia. In Europe and the United States, the strains isolated from clinical cases are mostly represented by CC1 strains accounting for 11.4% [5].
Until 1980, in Russia, lineage II strains isolated from human and animal listeriosis cases as well as from natural sources prevailed; most of them belonged to CC7 (also known as epidemic clone ECVII) [6]. CC7 strains still prevail in natural foci in the European part of Russia; they are also frequently isolated from sporadic cases of listeriosis in humans and animals [7]. Lineage II CC9 strains are often isolated from food and environmental sources; they are also well represented in Russia [7]. In the infectious disease pathology classification in Europe and North America, CC7 strains are assigned to medium virulent strains, while clonal complex CC9 is considered low virulent [8]. The difference in the occurrence frequency among clinical isolates and, consequently, the difference in the potential virulence of strains belonging to different CCs and/or phylogenetic lineages are associated by multiple researchers with the presence of additional virulence factors [9], allelic variation of virulence factors [10], and their expression levels [11].
The initial stage of infection starts with the interaction between Listeria and non-professional phagocytes, involving proteins belonging to the internalin family — InlA and InlB, which promote the L. monocytogenes invasion of intestinal enterocytes and epithelial cells through an interaction with target receptors. E-cadherin is a specific receptor for InlA. InlB specifically interacts with two receptors: c-Met and gC1q-R [12]. The presence of functional InlA is sufficient for invasion into enterocytes [13]; the entry into hepatocytes requires InlB [14], and the coordinated action of InlA and InlB mediates the crossing of the placental barrier [15].
Sequencing is performed to identify allelic variation of inlA and inlB genes. The study results show that clinical strains and, most importantly, strains causing fetal and neonatal infections express full-length InlA more often than strains of food origin. Truncated InlA was detected in most of the hypovirulent CC9 and CC121 isolates [16] and in other clonal complexes of phylogenetic lineage II (for example, CC331, CC199, and CC321) [4].
Earlier it was found that clinical and food isolates differed in inlA and inlB gene expression levels [11]. The expression of key pathogenicity factors is activated during cell infection and depends on transcriptional regulator PrfA. L. monocytogenes has an extensive cross-talk system between σB and PrfA regulatory networks, providing optimal expression of genes required in extrahost environments, including repression of genes associated with virulence. Conversely, in intrahost environments, this regulatory network provides elevated expression of genes associated with virulence, enabling successful invasion [17]. The PrfA activity decreases when L. monocytogenes is grown in rich media. Hydrophobic adsorbents included in the culture medium activate PrfA and induce enhanced activity of virulence factor genes [18]. However, the mechanisms regulating production levels of the respective proteins-pathogenicity factors remain unknown.
Various techniques are used to detect and measure levels of protein expression, including enzyme activity assays and immunoblotting. Enzyme activity measurement cannot be used if proteins are not biocatalysts. Immunoblotting is a labor-intensive process unsuitable for testing large sample sizes. A test system based on the enzyme-linked immunosorbent assay (ELISA) was developed as an alternative method for the qualitative and quantitative assessment of InlA and InlB. Using this ELISA test system, we assessed L. monocytogenes isolates from the collection of the Gamaleya Research Center of Epidemiology and Microbiology (Gamaleya RCEM) for InlA and InlB expression levels and their correlation with the phylogenetic status of the strains.
Materials and methods
Bacterial strains and bacteria culturing conditions
The study was performed using Listeria spp. strains from the collection of Gamaleya RCEM (Table 1). Microorganisms were grown in a BHI liquid medium (Becton Dickinson) at 37оC with constant shaking at 180 rpm. PrfA was activated by supplementing the BHI broth with 1% (w/v) Amberlite XAD 4 hydrophobic adsorbent (Sigma-Aldrich).
Table 1. Listeria spp. strains used in the study
Strain | Characteristics | Source | Reference |
L. monocytogenes | |||
EGDe | CC9 | Type strain | BIGSdb ID = 1 |
EGDeΔinlB | inlB deletion | Kindly provided by Prof. J. Vazquez-Boland | |
EGDeΔinlA | inlA deletion | ||
VIMHA004 | CC2 | Newborn | BIGSdb ID = 3449 |
VIMHA007 | CC2 | Newborn | BIGSdb ID = 3450 |
VIMHA006 | СС2 | Newborn | BIGSdb ID = 3606 |
VIMHA009 | CC1 | Newborn | BIGSdb ID = 3452 |
VIMHA011 | CC1 | Newborn | BIGSdb ID = 3454 |
VIMHA010 | CC1 | Newborn | BIGSdb ID = 3453 |
VIMHA012 | CC1 | Newborn | BIGSdb ID = 3607 |
VIMHA017 | CC1 | Newborn | BIGSdb ID = 3609 |
L.mo25 | CC1 | Chicken | BIGSdb ID = 79358 |
L.mo70 | CC8 | Chicken | BIGSdb ID = 78808 |
L.mo78 | CC37 | Chicken | BIGSdb ID = 79363 |
VIMPH006 | CC7 | Carrier | BIGSdb ID = 3464 |
L.mo71 | CC59 | Chicken | BIGSdb ID = 78809 |
GIMC2010:LmcUH8 | СС7 | Carrier | BIGSdb ID = 42978 |
GIMC2032:LmcINH-1 | СС7 | Carrier | BIGSdb ID = 45728 |
GIMC2007:LmcIH1_3 | СС7 | Carrier | BIGSdb ID = 42975 |
766 | СС7 | Swine | BIGSdb ID = 5803 |
VIMPR134 | CC7 | Rodent | BIGSdb ID = 3459 |
VIMPR422 | CC7 | Rodent | BIGSdb ID = 3460 |
VIMPA064 | CC7 | Newborn | BIGSdb ID = 3455 |
L.mo84 | СС37 | Meat | BIGSdb ID = 79367 |
L.mo49 | CC9 | Chicken | BIGSdb ID = 79359 |
L.mo129/3 | CC9 | Dairy products | BIGSdb ID = 78797 |
L.mo75 | CC9 | BIGSdb ID = 79362 | |
GIMC2035:Lmc7218 | CC9 | Fish | BIGSdb ID = 45731 |
GIMC2017: Lmc921 | CC9 | Meat | BIGSdb ID = 42987 |
L.mo98-20 | CC9 | Dairy products | BIGSdb ID = 78795 |
LO28 | CC9 | Type strain | BIGSdb ID = 3364 |
L. ivanovii | |||
ATCC 19119 | Type strain | American Type Culture Collection | |
PCR analysis
The collection-derived L. monocytogenes isolates were lysed with lysozyme at a concentration of 20 µg/ml for 1 hour at 37ºC; then, the lysates were treated with proteinase K at a concentration of 25 µg/ml at 56ºC for 1 hour. The obtained samples were boiled in a water bath for 10 min. The amplification of inlA and inlB genes was performed in a Tertsik thermal cycler (DNA-Technology) using TransStart Taq DNA Polymerase (TransGen): stage 1 (1 cycle): 94ºC — 4 min; stage 2 (30 cycles): 94ºC — 30 sec, 52ºC — 30 sec, 72ºC — 2 min; stage 3 (1 cycle): 72ºC — 10 min. Primers were selected using the Ugene v. 40.1 software and synthesized by the Syntol Company. For the inlA gene, the primers were as follows: InlA1 5’-ggttgaaaagtatactagtagc-3’; InlA2 5’-ggaagatattagcccaatttc-3’, for the InlB gene: InlBF 5’-gcttatggatcctataattcaaaagaag, InlBR 5’-gaaaagctgcagaaaatccgccttc. Gel electrophoresis was used for detecting amplification products. Target DNA fragments were purified using a Dia-gene kit (Dia-M).
Sequencing
Samples were prepared for sequencing following the recommendations of the Genome Shared Resource Center. The DNA sequencing was performed using an ABI PRISM BigDye Terminator v. 3.1 reagent kit; the reaction products were analyzed using an automated sequencer — Applied Biosystems 3730 DNA Analyzer. To identify gene sequences, the following primers were used: InlA1 5’-ggttgaaaagtatactagtagc-3’; InlA2 5’-ggaagatattagcccaatttc-3’; InlA-R 5’-cttcttttgaattataggatccataagc-3’, InlA3 5’-ccaatatcccgggaaaagctat-3’, 5’-gcttatggatcctataattcaaaagaag-3’, InlBR 5’-gaaaagctgcagaaaatccgccttc-3’ InlB1 5’-gaagcaggatcccggataactgcac-3’, InlB2 5’-atagcgggttaagttgactgc-3’. The nucleotide sequences were analyzed using the Ugene v. 40.1 software and the BigSdb-Pasteur database1.
Production of polyclonal monospecific antibodies against InlA and InlB
Rabbits were immunized with purified recombinant InlA and InlB proteins, as previously described [19]. The work with laboratory animals was performed in compliance with ethical principles. The research protocol was approved by the Ethics Committee of the Gamaleya Research Centre of Epidmiology and Microbiology (protocol No. 54, July 24, 2023).
The globulin fraction of hyperimmune sera was precipitated by adding a saturated (NH4)2SO4 solution (Ruskhim). The precipitate containing IgG was dissolved in 0.01 M Na-phosphate buffer (pH 6.5) and dialyzed against the same buffer. The obtained solution was passed through a DEAE Sephadex A-50 column and the IgG fraction was collected in the void volume. The affinity column was used to separate antibodies against the respective recombinant InlA and InlB proteins immobilized on an activated BrCN Sepharose (GE Healthcare). The fraction obtained from the DEAE chromatography was washed with 0.3 M NaCl and eluted with 4.5 M MgCl2 (Ruskhim). The obtained samples were dialyzed against phosphate-buffered saline (Sigma-Aldrich), pooled, mixed with glycerol to a concentration of 50% and stored at –20оC.
Western blot analysis of L. monocytogenes lysates
Cell wall proteins were obtained from an overnight culture grown in a BHI broth supplemented with 1% Amberlite XAD-4. The lysates were separated by SDS-PAGE in a 10% gel under Laemmli denaturing conditions and transferred onto a 0.45 μm pore-size nitrocellulose membrane (Bio-Rad). The nitrocellulose membranes were incubated with obtained antibodies against InlA or InlB, respectively, at 1:10000 dilution for 1 hour. Then, the membranes were washed 3 times with TTBS buffer (Bio-Rad) and anti-rabbit IgG secondary antibodies (at 1:20000 dilution) were added. The target InlA and InlB proteins were visualized on the membrane using the TMB substrate (Thermo Fisher Scientific). GAPDH antibodies were used to control protein loading (at 1:1000 dilution (Thermo Fisher Scientific).
Indirect enzyme-linked immunosorbent assay
Bacterial cells were grown in a BHI medium for 24 hours; then, they were centrifuged and washed three times with phosphate-buffered saline. The obtained samples reached an optical density of OD600 = 0.2. To verify that the number of cells has not changed, the cell cultures were seeded in an agar medium. The wells of a 96-well plate were coated with aliquots containing 100 μl of the respective tested L. monocytogenes strains. The plates were incubated at 37ºC for 1 hour; then, they were washed three times with Tris-buffered saline containing 0.05% Tween 20 (TTBS) (Ruskhim); the unoccupied sites in the wells were blocked by adding 200 μl of 2% bovine serum albumin (Dia-M) diluted in phosphate-buffered saline for 30 min. Upon completion of the incubation, the blocking buffer was removed and 100 μl of anti-InlB or anti-InlA antibodies were added at 1:4000 dilution in TTBS and incubated for 1 hour at room temperature with continuous shaking at 140 rpm. Then, the wells were washed three times with TTBS and 100 μl of secondary antibodies (goat anti-rabbit IgG antibodies conjugated to horseradish peroxidase, Bio-Rad), were added to TTBS. Then, the plates were washed 6 times with 250 µl of TTBS and developed by adding 100 µl of TMB substrate. To stop the reaction, 100 μl of 2 M H2SO4 (Ruskhim) was used. The optical density was measured at the wavelength of 450 nm using an iMark spectrophotometer (BioRad). The InlA and InlB concentration was measured using the calibration curve (Fig. 1).
Fig. 1. 96-well plate and test results.
Statistical analysis
All tests were repeated at least 3 times. The statistical analysis included one-way ANOVA and Tukey's test. Values of p < 0.05 were considered statistically significant.
Results
Analysis of the occurrence frequency of isolates deposited in the BigSdb-Pasteur database depending on the phylogenetic status and the region of isolation
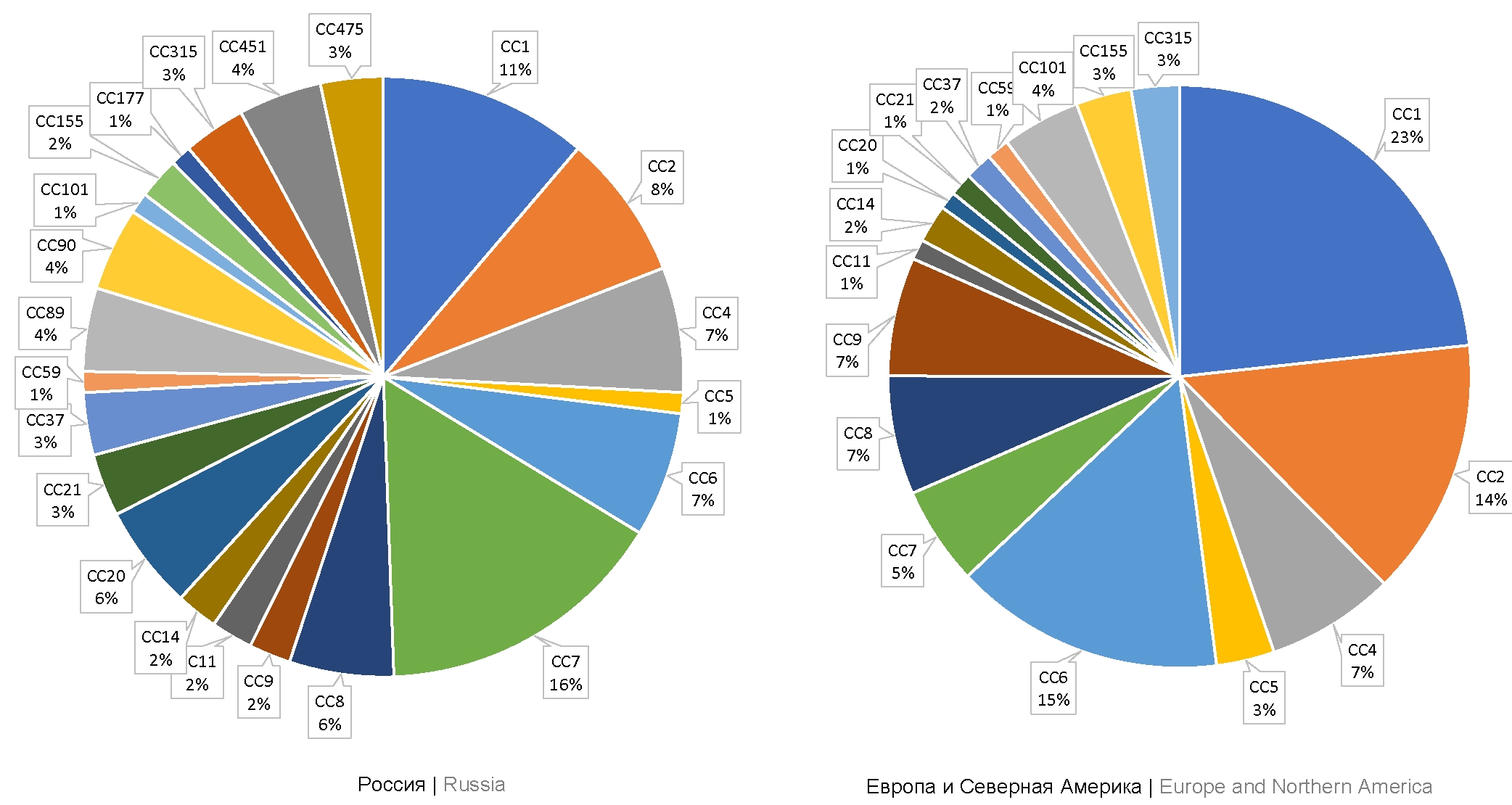
A total of 111 isolates collected in Russia from farm animals and humans infected with L. monocytogenes have been deposited in the BigSdb-Pasteur database. Out of them, 83 isolates were collected in 1958-2019 by researchers of the Somov Research Institute of Epidemiology and Microbiology, the Federal Research Center of Virology and Microbiology, Gamaleya RCEM and the State Scientific Center for Applied Microbiology and Biotechnology, using clinical materials collected from stillborn infants with perinatal listeriosis, from pregnant women without clinical signs of infection, and from samples collected from patients with neurolisteriosis and lung tissue lesions. The analysis of the database showed that human listeriosis cases were mostly caused by strains belonging to CC7 (phylogenetic lineage II), CC1, and CC2 (phylogenetic lineage I) accounting for 16, 11, and 8% of the isolates, respectively. Isolates belonging to CC9 (phylogenetic lineage II) accounted for 2% (Fig. 2).
Fig. 2. CC occurrence frequency among L. monocytogenes strains from the ListiList database (Institut Pasteur, France), for Russia, countries of Europe and North America.
Then, we analyzed the distribution with reference to CCs for isolates collected in Europe (excluding countries of the former Soviet Union) from clinical material of patients with listeriosis. A total of 747 isolates were deposited in the database. Note that 723 isolates represent the same clonal complexes that are identified in Russia. However, the percentage of some CCs among clinical isolates collected in Europe differed from those in Russia. Most of the European isolates belonged to CC1, CC6, and CC2 (23, 15, and 14%, respectively).
Assessment of the inlA and inlB diversity among L. monocytogenes isolates originated from Russia, Gamaleya RCEM collection
The database analysis showed that among phylogenetic lineage II strains prevailing in Russia, strains belonging to CC7 are more common among clinical isolates compared to strains belonging to CC9. To gain an insight into mechanisms responsible for these differences, we sequenced inlA and inlB genes of 32 strains belonging to the above CCs identified in Russia.
Among the strains belonging to CC7, we identified 3 inlA allelic variants; 15 strains had allele 2 of the inlA gene, which was presented in the BigSdb-Pasteur database. However, in 2 strains, inlA differed from the alleles deposited in the above database. The detected mutations were nonsynonymous, thus suggesting that substitutions may be of functional significance. The VIMPA064 isolate contained 1 nonsynonymous substitution leading to the substitution of glutamine at position 364 for proline in the internalin domain involved in the protein-protein interaction with the E-cadherin eukaryotic receptor. VIMPH006 contained 3 nonsynonymous substitutions at positions 618, 620, and 621 (proline, lysine, and glutamine instead of alanine, glutamine, and serine) outside the internalin domain. None of the alleles of CC7 strains contained premature stop codons.
Among 9 strains belonging to CC9, 1 strain had a stop codon in the inlA gene. The stop codon was located outside the internalin domain; however, it prevented the synthesis of the full-length protein and, specifically, the binding of the protein to the bacterial surface, which is essential for the functional activity of InlA as an invasion factor. The other 8 CC9 strains encoded the full-length inlA allele 1 variant. The data on sequences of inlA genes are available in the GenBank database under accession numbers OQ865090–OQ865119.
The inlB gene sequencing in 32 strains did not reveal any specific features within the clonal complexes. Allele 2 was identified for strains belonging to CC7, and allele 1 was identified for CC9 strains.
Thus, the analysis of inlA and inlB sequences showed that strains belonging to CC7 and CC9 were characterized by intraclonal variability of inlA, while inlB was conserved for all strains belonging to a specific clone. The alleles identified among highly virulent CC7 strains encoded different InlA isoforms, thus suggesting that there may be differences in their interaction with the target receptor on the surface of human cells. The variant encoded by the alternative inlA allele, which was identified among CC9 strains rarely found among clinical isolates, encoded a nonfunctional truncated protein.
Expression levels of the L. monocytogenes InlB pathogenicity factor in strains CC1, CC2, CC7, and CC9
Two ELISA-based test systems were designed to assess the correlation between the production levels of invasion factors InlA and InlB and the occurrence frequency of strains belonging to the above CCs among clinical isolates. The protein concentration was measured using calibration curves from 5 to 1000 ng/ml. Recombinant InlA and InlB were used as reference standards (Fig. 3).
Fig. 3. Immunoblot of Listeria spp. cell lysates. a — monospecific antibodies against InlA; b — monospecific antibodies against InlB. 1 – L. monocytogenes EGDe ΔinlA; 2 — L. ivanovii ATCC 19119; 3 — L. monocytogenes EGDe (CC9); 4 — L. monocytogenes EGDe ΔinlB; 5 — L. monocytogenes L.mo49 (CC9)
The PrfA regulator controls the transcription of both genes encoding InlA and InlB proteins [12]. When L. monocytogenes are grown in nutrient media, the PrfA activity increases in the presence of hydrophobic adsorbents (activated carbon or Amberlite XAD4), reaching levels typically demonstrated by bacteria during infection [18]. To analyze changes in the invasin levels depending on the PrfA status, we measured InlA and InlB levels without activation and with activation of the PrfA regulon, depending on the presence of the Amberlite XAD4 adsorbent in the medium. Without PrfA activation, the InlA level in L. monocytogenes strains ranged from 5 to 65 ng/ml (Fig. 4). On average, the strains belonging to CC1, CC2, and CC7 demonstrated higher InlA levels compared to the strains belonging to CC9 (53.5 ± 5.3 ng/ml compared to 21.1 ± 3.2 ng/ml; p < 0.05). The L.mo129-3 strain belonging to CC9 was an exception, demonstrating the level comparable with CC1, CC2, CC7 strains (59.1 ± 1.5 ng/ml).
Fig. 4. Levels of InlA isolates belonging to different CCs. The InlA concentration was measured using the new ELISA test system. Bacteria were grown in a BHI medium without and with the addition of 1% Amberlite XAD4.
The medium supplemented with the Amberlite XAD4 adsorbent and the resulting activation of PrfA led to increased InlA concentration in all strains by 31% on average. In some strains the concentration reached the highest levels: VIMHA017 (CC1) — 99 ng/ml, VIMHA007 (CC2) — 93 ng/ml, L.mo71 — 107 ng/ml (CC7), L.mo129-3 (CC9) — 90 ng/ml.
Among the strains, InlB levels ranged from 15 to 958 ng/ml without activation of the PrfA regulon (Fig. 5). On average, the concentration in CC1 and CC2 strains was 740.5 ng/ml, being significantly higher than in CC7 and CC9 strains (209.5 and 177.9 ng/ml, respectively). However, compared to InlA, the InlB level in L.mo129-3 strain did not reach values typical of CC1 and CC2 strains and was twice as low (p < 0.05).
Fig. 5. Levels of InlB isolates belonging to different CCs. The InlA concentration was measured using the new ELISA test system. Bacteria were grown in a BHI medium with and without 1% Amberlite XAD4.
When PrfA was activated, the InlB concentration reached the highest levels of 1,353 ng/ml for CC1 and CC2 strains, 917 ng/ml — for CC7 strains, 835 ng/ml — for CC9 strains. The highest InlB levels were demonstrated by CC7 strains (a 3.29-fold increase compared to the levels without activation). In the strains belonging to CC9 and phylogenetic lineage I (CC1, CC2), the InlB levels increased 2.93 and 1.54 times, respectively, in the presence of the adsorbent.
Discussion
In L. monocytogenes, like in most of the other pathogenic bacteria, clonal groups and lineages differ in their virulence potential [20, 21]. For example, phylogenetic lineage II strains are frequently associated with food products, while phylogenetic lineage I strains are commonly found in clinical isolates. However, the distribution among sources is different even within phylogenetic lineages. For example, phylogenetic lineage II strains belonging to CC7 are more frequently isolated from clinical samples than strains belonging to CC9 [20]. The underlying causes of this heterogeneity must be identified to understand the mechanisms of evolution and development of highly virulent strains.
In some pathogenic bacteria such as enteropathogenic Yersinia, the differences in virulence are associated with the presence of additional pathogenicity factors, which can be located on plasmids and mobile genetic elements [22]. The search of additional pathogenicity factors in L. monocytogenes strains belonging to clonal complex CC1 resulted in the discovery of LLS bacteriocin, which, presumably allows Listeria to survive longer in the presence of normal microbiota, though it is not a true pathogenicity factor and is absent in other highly virulent strains belonging to other CCs [23].
We have proposed an alternative hypothesis on the role of variability of major pathogenicity factors as the main mechanism responsible for development of highly virulent L. monocytogenes strains and other sapronotic pathogens [24]. Natural InlB isoforms differ from each other both in the kinetics of activation of intracellular signaling pathways and in the binding constant to their target receptors c-Met and gC1qR [25, 26]. Strains that differed only in the InlB isoform differed in their ability to support gastrointestinal infection in mouse-like rodents, while InlB isoforms were responsible for different invasion of these strains into animal epithelial cells (sheep kidney cells, shKEC) [10]. In our study, we demonstrated the variability of another invasion factor — the InlA protein among strains belonging to phylogenetic lineage II CCs: CC7 and CC9. Among the analyzed strains, we identified one strain with a nonfunctional InlA protein, thus demonstrating a relatively insignificant role of CC9 in the clinical spread of listeriosis and being consistent with the findings of other researchers [27].
The production level of major pathogenicity factors can be seen as another mechanism playing a significant role in the virulence potential of a strain [11]. It was previously found that the InlB production level affects the interaction of L. monocytogenes with macrophages [28]. In L. monocytogenes, the production of pathogenicity factors is controlled by the PrfA protein, which lacks activity in the growth environment outside the host (low temperature, presence of plant sugars and hydrophobic peptides) [22]. This regulatory mechanism is justified, as it eliminates the redundancy of the synthesis of pathogenicity factors for sapronotic pathogens, when they exist in the abiotic environment. At the same time, high production levels of factors required for crossing the intestinal epithelial barrier should have a positive effect on bacterial virulence. Following this hypothesis, this study demonstrated that the production level of the InlB invasion factor without PrfA activation was 4 times as high in phylogenetic lineage I strains (CC1 and CC2) compared to phylogenetic lineage II strains (CC7, CC9). The InlA production levels, without and with PrfA activation, in CC1, CC2, CC7 isolates were higher than the levels in CC9 isolates. This is consistent with the fact that CC1, CC2, CC7 are most often found in clinical samples in Russia. In CC9 strains, which are most common for food products, the InlA level was 2.5 times as low, except for the L.mo129-3 strain, which demonstrated levels similar to those observed in CC1 and CC2. Thus, our study has shown that strains highly virulent for humans are characterized not only by specific isoforms, but also by increased production of InlA and InlB. Our findings confirm the results obtained by other researchers who have found that clinical isolates differ from food isolates in inlA expression levels [11].
Conclusion
In total, our findings are consistent with the epidemiological monitoring data and demonstrate possible mechanisms of the formation of L. monocytogenes CCs with different virulence potentials. The obtained results are of fundamental importance; they can also serve as a basis for classifying newly collected isolates into virulent and hypovirulent clones.
1 URL: https://bigsdb.pasteur.fr/listeria
About the authors
Egor V. Kalinin
Gamaleya Research Center of Epidemiology and Microbiology
Email: kalinin.egor@bk.ru
ORCID iD: 0000-0002-4478-3225
researcher, Laboratory of ecology of infectious agents, Gamaleya Research Center of Epidemiology and Microbiology
Russian Federation, MoscowYaroslava M. Chalenko
Gamaleya Research Center of Epidemiology and Microbiology
Email: kalinin.egor@bk.ru
ORCID iD: 0000-0002-7901-5226
Cand. Sci. (Med.), senior researcher, Laboratory of ecology of infectious agents, Gamaleya Research Center of Epidemiology and Microbiology
Russian Federation, MoscowPolina V. Safarova
Gamaleya Research Center of Epidemiology and Microbiology
Email: kalinin.egor@bk.ru
ORCID iD: 0000-0001-9051-2664
Cand. Sci. (Med.), senior researcher, Laboratory of ecology of infectious agents, Gamaleya Research Center of Epidemiology and Microbiology
Russian Federation, MoscowVictoria A. Fedorova
Gamaleya Research Center of Epidemiology and Microbiology
Author for correspondence.
Email: kalinin.egor@bk.ru
ORCID iD: 0009-0000-9228-3818
laboratory researcher, Laboratory of ecology of infectious agents, Gamaleya Research Center of Epidemiology and Microbiology
Russian Federation, MoscowSvetlana A. Ermolaeva
Gamaleya Research Center of Epidemiology and Microbiology
Email: kalinin.egor@bk.ru
ORCID iD: 0000-0003-3396-6816
D. Sci. (Biol.), Head, Laboratory of ecology of infectious agents, Gamaleya Research Center of Epidemiology and Microbiology
Russian Federation, MoscowReferences
- Pires S.M., Desta B.N., Mughini-Gras L., et al. Burden of foodborne diseases: think global, act local. Curr. Opin. Food Sci. 2021;39:152–9. DOI: https://doi.org/10.1016/J.COFS.2021.01.006
- Halbedel S., Wilking H., Holzer A., et al. Large nationwide outbreak of invasive listeriosis associated with blood sausage, Germany, 2018-2019. Emerg. Infect. Dis. 2020;26(7):1456–64. DOI: https://doi.org/10.3201/eid2607.200225
- Rasmussen O.F., Skouboe P., Dons L., et al. Listeria monocytogenes exists in at least three evolutionary lines: еvidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology (Reading). 1995;141(Pt. 9):2053–61. DOI: https://doi.org/10.1099/13500872-141-9-2053
- Moura A., Criscuolo A., Pouseele H., et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016;2(2):1–10. DOI: https://doi.org/10.1038/nmicrobiol.2016.185
- Maury M.M., Tsai Y.H., Charlier C., et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016;48(3):308–13. DOI: https://doi.org/10.1038/ng.3501
- Psareva E.K., Egorova I.Y., Liskova E.A., et al. Retrospective study of Listeria monocytogenes isolated in the territory of inner Eurasia from 1947 to 1999. Pathogens. 2019;8(4):184. DOI: https://doi.org/10.3390/pathogens8040184
- Psareva E.K., Liskova E.A., Razheva I.V., et al. Diversity of Listeria monocytogenes strains isolated from food products in the central European part of Russia in 2000–2005 and 2019–2020. Foods. 2021;10(11):2790. DOI: https://doi.org/10.3390/foods10112790
- Charlier C., Perrodeau É., Leclercq A., et al. Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect. Dis. 2017;17(5):510–9. DOI: https://doi.org/10.1016/S1473-3099(16)30521-7
- Cotter P.D., Draper L.A., Lawton E.M., et al. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4(9):e1000144. DOI: https://doi.org/10.1371/journal.ppat.1000144
- Chalenko Y., Kolbasova O., Pivova E., et al. Listeria monocytogenes invasion into sheep kidney epithelial cells depends on InlB, and invasion efficiency is modulated by phylogenetically defined InlB isoforms. Front. Microbiol. 2022;13:825076. DOI: https://doi.org/10.3389/fmicb.2022.825076
- Severino P., Dussurget O., Vêncio R.Z.N., et al. Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall, and stress response. Appl. Environ. Microbiol. 2007;73(19):6078–88. DOI: https://doi.org/10.1128/AEM.02730-06
- Kammoun H., Kim M., Hafner L., et al. Listeriosis, a model infection to study host-pathogen interactions in vivo. Curr. Opin. Microbiol. 2022;66:11–20. DOI: https://doi.org/10.1016/j.mib.2021.11.015
- Lecuit M., Ohayon H., Braun L., et al. Internalin of Listeria monocytogenes with an intact leucine-rich repeat region is sufficient to promote internalization. Infect. Immun. 1997;65(12): 5309–19. DOI: https://doi.org/10.1128/iai.65.12.5309-5319.1997
- Dramsi S., Biswas I., Maguin E., et al. Entry of Listeria monocytogenes into hepatocytes requires expression of InIB, a surface protein of the internalin multigene family. Mol. Microbiol. 1995;16(2):251–61. DOI: https://doi.org/10.1111/j.1365-2958.1995.tb02297.x
- Disson O., Grayo S., Huillet E., et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature. 2008;455(7216):1114–8. DOI: https://doi.org/10.1038/nature07303
- Gelbíčová T., Koláčková I., Pantůček R., Karpíšková R. A novel mutation leading to a premature stop codon in inlA of Listeria monocytogenes isolated from neonatal listeriosis. New Microbiol. 2015;38(2):293–6.
- Gaballa A., Guariglia-Oropeza V., Wiedmann M., et al. Cross talk between SigB and PrfA in Listeria monocytogenes facilitates transitions between extra- and intracellular environments. Microbiol. Mol. Biol. Rev. 2019;83(4):e00034-19. DOI: https://doi.org/10.1128/MMBR.00034-19
- Ermolaeva S., Novella S., Vega Y., et al. Negative control of Listeria monocytogenes virulence genes by a diffusible autorepressor. Mol. Microbiol. 2004;52(2):601–11. DOI: https://doi.org/10.1111/j.1365-2958.2004.04003.x
- Kalinin E.V., Chalenko Y.M., Kezimana P., et al. Combination of growth conditions and InlB-specific dot-immunoassay for rapid detection of Listeria monocytogenes in raw milk. J. Dairy Sci. 2023;106(3):1638–49. DOI: https://doi.org/10.3168/JDS.2022-21997
- Maury M.M., Bracq-Dieye H., Huang L., et al. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019;10(1):1–13. DOI: https://doi.org/10.1038/s41467-019-10380-0
- Quereda J.J., Morón-García A., Palacios-Gorba C., et al. Pathogenicity and virulence of Listeria monocytogenes: а trip from environmental to medical microbiology. Virulence. 2021;12(1):2509–45. DOI: https://doi.org/10.1080/21505594.2021.1975526
- Timchenko N.F., Adgamov R.R., Popov A.F., et al. Far Еast scarlet-like fever caused by a few related genotypes of Yersinia pseudotuberculosis, Russia. Emerg. Infect. Dis. 2016;22(3): 503–6. DOI: https://doi.org/10.3201/EID2203.150552
- Quereda J.J., Andersson C., Cossart P., et al. Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet. Res. 2018;49(1):1–9. DOI: https://doi.org/10.1186/S13567-017-0496-4
- Ермолаева С.А., Зайцева Е.А., Тимченко Н.Ф., Адгамов Р.Р. Вариабельность функциональных доменов факторов инвазии как молекулярная основа полигостальности возбудителей сапронозов. Тихоокеанский медицинский журнал 2010;(4):24–8. Ermolaeva S.A., Zaitseva E.A., Timchenko N.F., Adgamov R.R. Variability of functional domains of invasion factors as molecular basis for polyhostality of sapronosis-induced microorganisms. Pacific Medical Journal. 2010;(4):24–8. EDN: https://elibrary.ru/ocqdnb
- Chalenko Y.M., Kalinin E.V., Marchenkov V.V., et al. Phylogenetically defined isoforms of Listeria monocytogenes invasion factor InlB differently activate intracellular signaling pathways and interact with the receptor gC1q-R. Int. J. Mol. Sci. 2019;20(17):4138. DOI: https://doi.org/10.3390/ijms20174138
- Chalenko Y.M., Slonova D.A., Kechko O.I., et al. Natural isoforms of Listeria monocytogenes virulence factor InlB differ in c-Met binding efficiency and differently affect uptake and survival listeria in macrophage. Int. J. Mol. Sci. 2023;24(8):7256. DOI: https://doi.org/10.3390/ijms24087256
- Phelps C.C., Vadia S., Arnett E., et al. Relative roles of listeriolysin O, InlA, and InlB in Listeria monocytogenes uptake by host cells. Infect. Immun. 2018;86(10):e00555-18. DOI: https://doi.org/10.1128/iai.00555-18
- Чаленко Я.М., Абдулкадиева М.М., Сафарова П.В. и др. Белок INLB, секретируемый Listeria monocytogenes, контролирует взаимодействие возбудителя с макрофагами. Вестник Российского государственного медицинского университета. 2022;(3):5–10. DOI: https://doi.org/10.24075/vrgmu.2022.034 Chalenko Y.M., Abdulkadieva M.M., Safarova P.V., et al. InlB protein secreted by Listeria monocytogenes controls the pathogen interaction with macrophages. Bulletin of Russian State Medical University. 2022;(3):5–10. DOI: https://doi.org/10.24075/brsmu.2022.034
Supplementary files