Bacteria of genus Filifactor in patients with periodontitis and type 2 diabetes in accordance with metagenomic analysis of the periodontal microbiome
- Authors: Tsareva T.V.1, Yanushevich O.O.1, Tsarev V.N.1, Balmasova I.P.1
-
Affiliations:
- Moscow State University of Medicine and Dentistry named after A.I. Evdokimov
- Issue: Vol 100, No 6 (2023)
- Pages: 485-494
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/16885
- DOI: https://doi.org/10.36233/0372-9311-428
- EDN: https://elibrary.ru/yqoxxk
- ID: 16885
Cite item
Abstract
Introduction. Periodontal diseases are a common pathology with chronic periodontitis (CP) being the most severe form. This polymicrobial disease has become a problem of great importance in recent years due to the possibility of development of systemic effects associated with this condition. CP is often combined with type 2 diabetes (T2D). The main cause of the occurrence and development of periodontal pathology is played by the bacteria Filifactor alocis, the least studied and most recently discovered periodontal pathogen.
The objective of this study was to identify bacteria of genus Filifactor as part of the periodontal microbiome associated with CP and T2D and to clarify the mechanisms of their possible influence on associated metabolic processes according to comparative metagenomic analysis.
Materials and methods. A metagenomic study of the microbiome of periodontal pocket samples from 28 patients with CP associated with T2D and 22 patients with CP, as well as the microbiome of dental gingival sulcus samples from 19 clinically healthy individuals was performed. 16S-sequencing of the ribosomal RNA gene was used to determine the taxonomic composition of the microbiome. Prediction of metabolic pathways involving the microbiome was performed with the help of the shotgun method.
Results. Filifactor bacteria were the one of the most frequent microorganisms only in patients with CP associated with T2D. The rate of identification of these bacteria was correlated with low predicted metagenomic levels of fatty acid biosynthesis and pyrimidine metabolism in the affected area.
Conclusion. The detection frequency of Filifactor bacteria in patients associated with CP and T2D is negatively correlated with the selected features of putative metabolic pathways of the microbiome, which include fatty acid biosynthesis and pyrimidine metabolism.
Full Text
Introduction
Periodontal diseases are widespread among the adult population of our planet. Thus, about 740 million people in the world suffer from severe periodontitis [1]. The relevance of this problem is even greater due to the fact that in recent years both pathogenetic and causal relationship has been established between oral diseases and somatic diseases [2]. The relationship between periodontitis and such systemic pathologic conditions as type 2 diabetes (T2D), rheumatoid arthritis, atherosclerosis, adverse pregnancy outcomes, etc. is widely discussed in other studies [3–5]. The persistent association of T2D and chronic periodontitis (CP) as the two diseases with the highest prevalence worldwide is of particular importance [6, 7].
The International Diabetes Federation predicts that the incidence of diabetes will steadily increase over a long period of time and the number of patients will exceed 500 million people by 2030 [8, 9]. Among patients with diabetes, about 90% are patients with T2D, a non-insulin-dependent diabetes caused by a combination of decreased insulin secretion by pancreatic β-cells and increased cellular resistance to insulin [10]. Many researchers show that metabolic changes associated with diabetes are manifested at the systemic level and contribute to the damage of many organs and tissues, including the development of CP [11, 12], although the mutual influence of these pathological processes is not excluded either [3, 5].
CP has a polymicrobial nature. It is caused by a number of microorganisms with properties of periodontal pathogens and characterized by irreversible course, accompanied by chronic inflammation of periodontal tissues and its destruction along with damage to the bone structure and tooth supporting apparatus, which eventually leads to tooth loss. All the main causative agents of this disease (Porphyromonas gingivalis, Tannerella forsythia, Aggregatibacter actinomycetemcomitans, Filifactor alocis, Porphyromonas endodontalis, Treponema denticola, Prevotella intermedia, Fusobacterium nucleatum, etc.), the so-called periodontal pathogens of the I and II order or bacteria of the "red" and "orange" complexes, as they were originally called in international literature, are in complex interactions with each other and with the immune system of the host organism, which provokes both the development of local changes in periodontal tissues and the following consequences of these inflammatory and degenerative changes on the systemic level [3, 13, 14].
Thanks to the development of biotechnology, there is enough evidence that the oral microbiome in CP is the main factor in the development of diabetes [15], although there are still uncertainties in the study of the role of individual periodontal pathogens in this problem.
Most studies on the relationship between periodontal and systemic diseases have focused primarily on the role of Porphyromonas gingivalis, which is considered a key periodontal pathogen [3, 14]. In recent years, emphasis has been placed on the study of other red complex periodontal pathogens. In addition to the red complex bacteria, attention has been called upon the importance of other culturable bacteria (Prevotella intermedia, Fusobacterium nucleatum, Eykinella corrodens, Eubacterium nodatum, Wolinella recta) associated with periodontitis, as well as non-culturable (under the conditions of a conventional clinical laboratory) bacteria — Selenomonas, Sinergistes, Desulfobulbus, TM7, Filifactor alocis, identified as new potential pathogens or orange complex periodontal pathogens [3, 13, 16, 17].
One of the representatives of these species, Filifactor alocis (F. alocis), is a Gram-positive spore-forming bacillus with obligatory anaerobic respiration, forming filamentous clusters in the gingival biofilm, which grows extremely slowly on nutrient media and has low biochemical activity, which makes its identification difficult [18, 19]. F. alocis is an asaccharolytic bacterium that utilizes certain amino acids, especially arginine, for its growth [20].
The main habitat of F. alocis in the human body is the gingival sulcus and possibly the intestine, while a healthy person does not usually contain these bacteria in the microbiome of their oral cavity (except for smokers), unlike people suffering from periodontal disease [20]. F. alocis bacteria have unique properties such as resistance to oxidative stress, the ability to induce systemic bone mass loss, the presence of genes encoding a well-developed amino acid metabolism pathway, which allows them to colonize periodontal tissues, to cause periodontitis phenomena in the stressful environment of the forming periodontal pocket like other traditional periodontal pathogens [21-23]. These features of F. alocis, in addition to its ability to interact with other microbial species to form polymicrobial synergistic relationships, may enhance the invasive properties of these bacteria [24] and cause chronic inflammation [25]. F. alocis can penetrate gingival epithelial cells and induce their secretion of pro-inflammatory cytokines, which contributes to the manifestations of pathogenicity of this microbe [26].
Taken together, all these observations point to a specific role of F. alocis in the oral cavity, which may be important in the pathologic process [27, 28]. Due to the lack of genetic tools to study this microorganism, for a long time little was known about its ability to influence microbial metabolism in biofilm, its mechanisms of virulence and especially, its role in the induction of systemic effects [21].
Accordingly, the objective of the present study was to identify Filifactor bacteria in the periodontal microbiome with the association of CP and T2D and to clarify the mechanisms of their possible influence on the associated metabolic processes based on metagenomic analysis.
Materials and methods
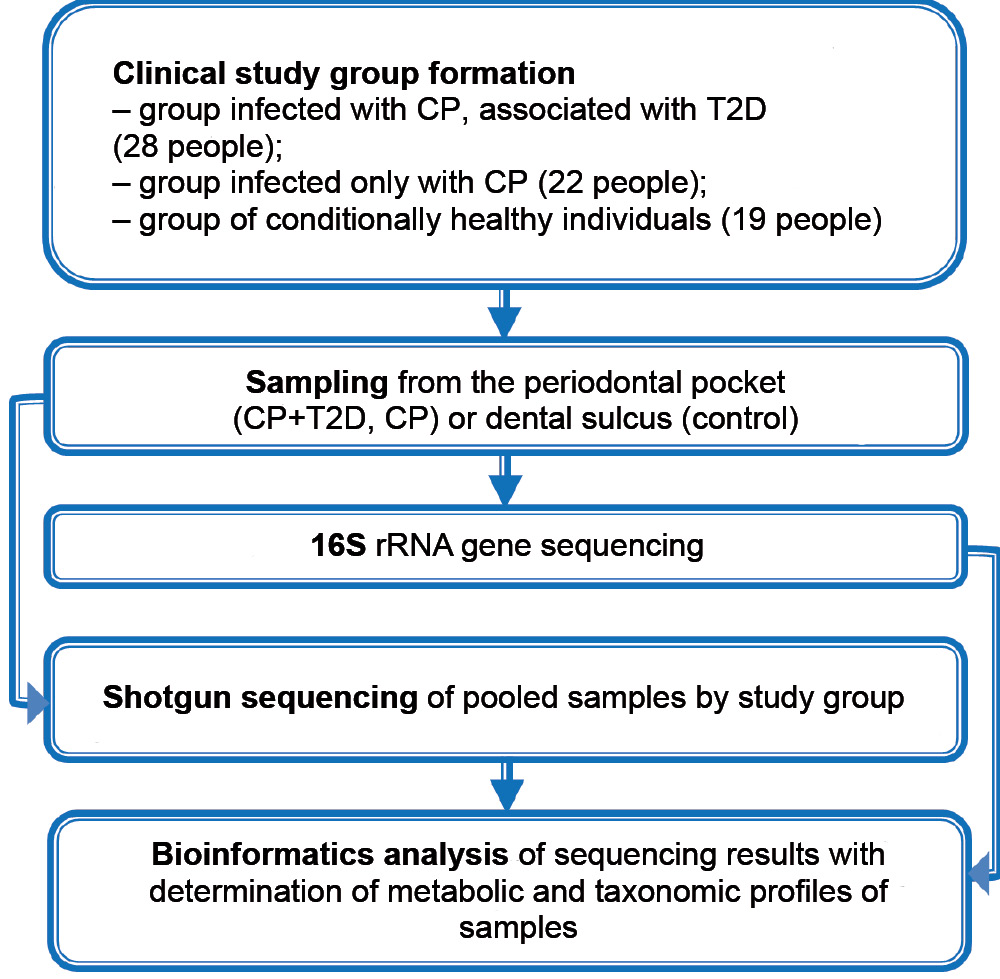
Sixty-nine people aged 40-65 years were under observation. The main group included 28 patients with the association of CP and T2D; the comparison group consisted of 22 patients with CP without concomitant somatic pathology; the control group included 19 conditionally healthy individuals with intact periodontium, without clinically expressed manifestations of somatic pathology, with normal blood sugar and glycated hemoglobin levels (Fig. 1). The number of women and men in the comparison groups was nearly equal: 13 (46.5%) and 15 (53.5%) in the CP + T2D group, 10 (45.4%) and 12 (54.6%) in the CP group, 9 (47.4%) and 10 (2.6%) in the control group.
Due to the necessity of using pooled samples of biological material for metagenomic studies, the maximum homogeneity of the study groups was a requirement; therefore, patients with mild and severe course of pathological processes were excluded from the study, in other words, all patients under study had a moderately severe course of the diseases. Patients did not receive treatment for CP during the last 6 months. Among patients with T2D, 11 patients received insulin therapy, 13 patients — oral sugar-lowering drugs, 4 patients — combined therapy, which was taken into account when distributing the samples for further pooling.
CP was diagnosed based on clinical and radiologic findings according to the 2018 classification of periodontal diseases [29]. Inclusion criteria of these patients in the study were as follows: periodontitis of medium severity with generalized lesions, the value of loss of gingival attachment CAL 3–4 mm, the depth of probing pockets 4–6 mm, bone loss around teeth not more than 1/3 of the root length, absence of tooth loss associated with the disease. To confirm the diagnosis, we analyzed anamnesis data, determined the PHR (Patient Hygiene Performance), OHI-S (Simplified Oral Hygiene Index), CAL (Clinical Attachment Level), MMI (Miller Mobility Index), PBI (Papillary Bleeding Index), TL (tooth loss) and the orthopantomography (OPG) data.
The diagnosis of T2D was established by an endocrinologist in accordance with the diagnostic criteria of the World Health Organization 1999/2006/2011 [30], taking into account the clinical, anamnestic and laboratory data. Inclusion criteria in the study were as follows: disease duration 3–7 years; moderately compensated disease course; remission stage; blood glucose level < 7.8 mmol/l, glycated hemoglobin level < 8%.
Conditionally healthy control subjects were included in the study only after consultation with a dentist and endocrinologist.
The material for the study was the content of periodontal pockets from patients with CP and dental gingival sulcus in healthy subjects. The material was taken from 4 sites in the area of the gingival sulcus using sterile paper endodontic pins (#30), which were placed in a test tube with 0.2 ml of a sterile physiologic solution and shaken. The samples were stored at –20ºC. The study was conducted with the informed consent of the patients. The research protocol was approved by the Ethics Committee of the Interuniversity Ethics Committee of Moscow (protocol No. 06-22, June 6, 2022).
Total DNA was extracted from samples using the QIAamp DNA Investigator Kit (Qiagen), the concentration of genomic DNA was determined on a Qubit 2.0 fluorometer (Invitrogen) according to the manufacturer's instructions. Enriched microbial DNA (50–100 ng) was fragmented using a Covaris S220 system (Covaris). The final fragment size was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies) according to the manufacturer's instructions. The purified DNA was amplified using standard primers for the 16S rRNA gene complementary to the V3–V4 region and containing 5'-illumina adaptor sequences.
Sequencing of libraries and analysis of obtained data were performed using MiSeq genetic analyzer (Illumina) and MiSeq Reagent Kit v2 (Pharma).
The microbiome bioinformatics platform QIIME2 (Quantitative Insights into Microbial Ecology) [31] and the SILVA database [32] were used to perform taxonomic analysis of the results of sequencing of variable regions of the 16S rRNA gene. Identification of differences at the level of taxonomic compositions was performed by variance analysis of data based on the PERMANOVA test for microbial communities from the QIIME2 functional [33]. The PICRUST2 (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) method [34] was used to predict metabolic profile from sequencing and 16S rRNA gene profiling data [34], and STAMP statistical analysis of metabolic profiles was used to detect statistical differences between groups at the level of predicted metabolic pathways [35]. Data of metagenomics analysis were visualized using GNU/R software and Vegan package [36].
Results
At the first stage, 16S gene sequencing of the small subunit of ribosomal RNA was performed, which allowed us to determine the representation of different taxonomic categories of bacteria in the samples from the contents of the patients' dental sulcus.
The bioinformatic analysis revealed quite significant features of the microbial composition of the dental sulcus contents in CP associated with T2D which have never before been noted by other researchers. The features are as follows: significant predominance of 5 taxonomic groups of bacteria — representatives of the Filifactor and Mycoplasma genera, Flavobacteriaceae and Porphyromonadaceae families, Bacillales order —and lower occurrence of 3 groups: TM7 as associates of the actinomycetes, Atopobium and Fusobacterium genera (Fig. 2).
Fig. 2. Taxonomic profile of periodontal pocket microbiome in comparison of study groups.
The comparative analysis of the taxonomic profile of microbiome of patients with CP, with or without association with T2D, showed more frequent occurrence in the first group of bacteria of the Filifactor and Treponema genera and lower frequency of occurrence in the microbiome of 5 taxonomic groups: Bordetella and Atopobium genera, Fusobacteriaceae and Veillonellaceae families, TM7.
In patients with CP without somatic pathology in comparison with healthy people, bacteria of 5 taxonomic groups: Anaerosinus and Bulleidia genera, as well as the Bacillales, Bacteroidales, and Macellibacteroides orders were encountered more frequently.
In general, the main peculiarity of the taxonomic profile of the gingival sulcus microbiome with the association of CP+T2D is the predominance of bacteria with periodontal pathogenic properties belonging to the Filifactor and Treponema genera in the samples. The more frequent occurrence of prokaryotes of the Mycoplasma genus, Flavobacteriaceae family and Bacillales order compared to healthy people is a topic of interest and requires further analysis.
One important fact that should be taken into account is that the most significant feature of the group of patients with CP+T2D is the predominant representation of Filifactor genus bacteria in the microbiome. According to this feature, the microbiome of this group differs both from the group of healthy people and from the group of patients with only moderate CP, in which Filifactor genus bacteria are found in isolated cases. The question that this brings up is to what extent this feature is associated with the development of diabetes and those metabolic changes observed in T2D and associated mainly with the effect of the microbiome.
At the next stage, sequencing of pooled samples was performed to determine the quantitative prevalence of certain taxonomic categories of bacteria in the microbiome by individual pools of biological material with functional traits that allowed to predict the main metabolic pathways of the analyzed pooled microbiome samples belonging to different clinical groups (15 pooled samples in each group).
Differences in the taxonomic diversity of metagenomes were fully reflected in the level of differences in the predicted functional potential of microbial communities by study groups (Fig. 3).
Fig. 3. Predicted profile of periodontal pocket microbiome metabolic pathways in the comparison of the clinical study groups.
A decrease in relative representation of 8 metabolic pathways was found in the CP+T2D group compared to the controls: fatty acid biosynthesis, pyrimidine metabolism, methane metabolism, glycerolipid metabolism, sphingolipid metabolism, tyrosine metabolism, ascorbate and aldarate metabolism, ubiquinone and other terpenoid-quinones biosynthesis.
When comparing the groups of CP with and without T2D, differences in the representation of 5 metabolic pathways were revealed: in the group of CP+T2D, an increase in cysteine and methionine metabolism and a decrease in pyrimidine metabolism, fatty acid biosynthesis, methane metabolism, and sphingolipid metabolism were observed.
In comparison to the controls, a decrease in the relative representation of 4 metabolic pathways: cysteine and methionine, sulfur, histidine, glycerolipids was found for the CP group.
Ultimately, 4 metabolic pathways can be identified, the low level of which distinguishes the CP+T2D group from controls and the CP group: methane metabolism, sphingolipid metabolism, fatty acid biosynthesis, and pyrimidine metabolism. CP, regardless of association with T2D, was characterized by decreased glycerolipid metabolism.
Functional analysis of pooled samples using the Bray–Curtis criterion confirmed that, in general, the group of CP patients was significantly closer to the control in characterizing metabolic pathways than the CP+T2D group.
Correlation analysis using Spearman correlation coefficient (r) was performed to determine the relationship between the revealed features of metabolic profiles by study groups and the presence of representatives of the genus Filifactor in the composition of the periodontal pockets microbiome. The frequency of occurrence of these bacteria in CP+T2D in our study showed a significant level of negative correlation (r > 0.723) with the prediction of two metabolic pathways — fatty acid biosynthesis and pyrimidine metabolism, while all these features in patients of this group were registered 3-5 times less frequently than in other cases.
Discussion
The highest occurrence of Filifactor bacteria, relatively recently classified to the group of periodontal pathogens of the first order [19], in the association of CP+T2D has been found by other authors among the predominant representatives of the microbiome of periodontal pockets [37–40], although without a detailed comparison of the occurrence of this microorganism in other groups.
The data obtained by us on the peculiarities of periodontal pockets microbiome metabolism in patients with CP+T2D indicate a rather significant role of disorders of fatty acids, pyrimidine, methane, glycerolipids, and sphingolipids metabolism. Two of these features (decreased biosynthesis of fatty acids and pyrimidine metabolism) were correlated with the presence of Filifactor bacteria in the microbiome. The fact of such correlation was confirmed by us for the first time; at the same time, the importance of these metabolic pathways of the microbiome with the association of CP+T2D is discussed in modern scientific literature.
For example, when evaluating the metabolism of F. alocis, there is emphasis on the importance of the biosynthesis of saturated fatty acid, butyric acid, which is involved in the pathogenesis of periodontitis [41]. Moreover, it is even proposed to use microbial butyric acid as a marker of periodontitis, emphasizing that this compound may affect the body's sensitivity to insulin [42]. There is an opinion that the oral microbiome in CP+T2D may be characterized by a decrease in the number of bacteria producing butyric acid [43]. This somewhat contradicts the data of our study, at least in terms of significant representation of the Filifactor genus in the microbiome of the pathology under consideration. At the same time, the deficiency of fatty acids that have been established with the association of CP+T2D may have another explanation. For example, if herpesviruses (especially Epstein-Barr virus) are present in the microbiome of subgingival biofilm, they have the ability to actively consume the same butyric acid for their reactivation [44]. All of this shows the possibility of ambiguous interpretation of the obtained results and the necessity for further research in this area.
The decrease in pyrimidine metabolism, correlated with the occurrence of F. alocis, has also been reported in the scientific literature as a sign of CP. For example, there are reports that genes involved in pyrimidine synthesis had significantly lower relative abundance in patients with periodontitis when compared to healthy controls [45]. Pyrimidine is an agonist of the GR119 receptor associated with hypoglycemic effects and a protective effect against pancreatic β-cells, which even allows us to recommend pyrimidine derivatives as therapeutic agents in T2D [46]. From this point of view, the state of pyrimidine metabolism in the microbiome of periodontal tissues with the participation of F. alocis may prove to be a kind of marker of the association of periodontitis with diabetes mellitus, and possibly one of the numerous elements of T2D pathogenesis.
Thus, further study of these phenomena is promising not only from the point of view of deciphering the etiopathogenetic role of bacteria of the genus Filifactor, but also from the point of view of improving the methods of diagnostics and treatment of the above mentioned combined pathology.
Conclusion
- The microbiome of periodontal tissues in patients with CP associated with T2D has a number of peculiarities concerning both its functional features and taxonomic composition. In the latter case, researchers concentrate their focus on recently established and so far poorly studied bacterial representatives of the microbiome that are unculturable and have pronounced periodontal pathogenic properties. Such microorganisms include bacteria of F. alocis species in particular.
- During a metagenomic microbiome analysis of periodontal pockets of patients, Filifactor bacteria were found to be significantly more frequent in the microbiome of patients with CP+T2D than in those who only have periodontitis or in healthy individuals. The detection rate of Filifactor bacteria in patients with CP+T2D was negatively correlated with certain features of the presumed metabolic pathways of the microbiome, which included fatty acid biosynthesis and pyrimidine metabolism.
- The noted metabolic features, in particular, a decrease in pyrimidine metabolism correlating with the occurrence of F. alocis, according to the literature, with a certain degree of probability can be considered a pathogenetic factor in the development of periodontitis and/or T2D and can be regarded as a diagnostic marker.
About the authors
Tatyana V. Tsareva
Moscow State University of Medicine and Dentistry named after A.I. Evdokimov
Email: tancha-leo84@mail.ru
ORCID iD: 0000-0001-9571-0520
SPIN-code: 2028-8404
Scopus Author ID: 39762981000
https://www.msmsu.ru/obrazovanie/teaching-staff/2952/
Cand. Sci. (Med.), Associate Professor, Department of microbiology, virology, immunology
Russian Federation, MoscowOleg O. Yanushevich
Moscow State University of Medicine and Dentistry named after A.I. Evdokimov
Email: msmsu@msmsu.ru
ORCID iD: 0000-0003-0059-4980
SPIN-code: 1452-1387
Scopus Author ID: 57371027600
https://www.msmsu.ru/university/rektorat/12/
D. Sci. (Med.), Professor, RAS Full Member, Head, Department of periodontology, Rector
Russian Federation, MoscowViktor N. Tsarev
Moscow State University of Medicine and Dentistry named after A.I. Evdokimov
Email: nikola777@rambler.ru
ORCID iD: 0000-0002-3311-0367
SPIN-code: 8180-4941
Scopus Author ID: 7005569282
https://www.msmsu.ru/obrazovanie/teaching-staff/1303/
D. Sci. (Med.), Professor, Head, Department of microbiology, virology, immunology
Russian Federation, MoscowIrina P. Balmasova
Moscow State University of Medicine and Dentistry named after A.I. Evdokimov
Author for correspondence.
Email: iri.balm@mail.ru
ORCID iD: 0000-0001-8194-2419
SPIN-code: 8025-8611
https://www.msmsu.ru/obrazovanie/teaching-staff/6807/
D. Sci. (Med.), Professor, Head, Laboratory of pathogenesis and methods of treatment of infectious diseases
Russian Federation, MoscowReferences
- Eke P.I., Wei L., Borgnakke W.S., et al. Periodontitis prevalence in adults ≥65years of age, in the USA. Periodontol. 2000. 2016;72(1):76–95. DOI: https://doi.org/10.1111/prd.12145
- Мамедов М.Н., Куценко В.А., Керимов Э.Ф. и др. Связь состояния соматического здоровья с заболеваниями тканей пародонта и зубов в реальной клинической практике. Профилактическая медицина. 2022;25(2):66–73. Mamedov M.N., Kutsenko V.A., Kerimov E.F., et al. Relationship between the state of somatic health and diseases of periodontal tissues and teeth in real clinical practice. The Russian Journal of Preventive Medicine. 2022;25(2):66–73. DOI: https://doi.org/10.17116/profmed20222502166 EDN: https://elibrary.ru/xckwlj
- Балмасова И.П., Царев В.Н., Янушевич О.О. и др. Микроэкология пародонта. Взаимосвязь локальных и системных эффектов. М.; 2021:175–225. Balmasova I.P., Tsarev V.N., Yanushevich O.O., et al. Microecology of Periodontium. The Relationship Between Local and Systemic Effects. Moscow; 2021:175–225. EDN: https://elibrary.ru/myzmbu
- Hajishengallis G., Chavakis T. Local and systemic mechanisms linking periodontal disease and inflammatory comorbidities. Nat. Rev. Immunol. 2021;21(7):426–40. DOI: https://doi.org/10.1038/s41577-020-00488-6
- Nikolaeva E.N., Tsarev V.N., Tsareva T.V., et al. Interrelation of cardiovascular diseases with anaerobic bacteria of subgingival biofilm. Contemp. Clin. Dent. 2019;10(4):637–42. DOI: https://doi.org/10.4103/ccd.ccd_84_19
- Bascones-Martínez A., Muñoz-Corcuera M., Bascones-Ilundain J. Diabetes and periodontitis: A bidirectional relationship. Med. Clin. (Barc.). 2015;145(1):31–5. DOI: https://doi.org/10.1016/j.medcli.2014.07.019 (in Spanish)
- Portes J., Bullón B., Quiles J.L., et al. Diabetes mellitus and periodontitis share intracellular disorders as the main meeting point. Cells. 2021;10(9):2411. DOI: https://doi.org/10.3390/cells10092411
- Alam U., Asghar O., Azmi S., Malik R.A. General aspects of diabetes mellitus. Handb. Clin. Neurol. 2014;126:211–22. DOI: https://doi.org/10.1016/b978-0-444-53480-4.00015-1
- Ida S., Kaneko R., Murata K. Utility of real-time and retrospective continuous glucose monitoring in patients with type 2 diabetes mellitus: A meta-analysis of randomized controlled trials. J. Diabetes Res. 2019;2019:4684815. DOI: https://doi.org/10.1155/2019/4684815
- American Diabetes Association. Classification and diagnosis of diabetes: standards of medical care in diabetes – 2021. Diabetes Care. 2021;44(Suppl. 1):15–33. DOI: https://doi.org/10.2337/dc21-s002
- Patil V.S., Patil V.P., Gokhale N., et al. Chronic periodontitis in type 2 diabetes mellitus: oxidative stress as a common factor in periodontal tissue injury. J. Clin. Diagn. Res. 2016;10(4):BC12–6. DOI: https://doi.org/10.7860/jcdr/2016/17350.7542
- Унаньян К.Г., Балмасова И.П., Царев В.Н. и др. Церамиды как биомаркеры хронического пародонтита, ассоциированного с сахарным диабетом второго типа. Вестник Российского университета дружбы народов. Серия: Медицина. 2020;24(4):325–37. Unanyan K.G., Balmasova I.P., Tsarev V.N., et al. Ceramides as biomarkers of chronic periodontitis associated with type II diabetes. RUDN Journal of Medicine. Series: Medicine. 2020;24(4):325–37. DOI: https://doi.org/10.22363/2313-0245-2020-24-4-325-337 EDN: https://elibrary.ru/kykdxt
- Царев В.Н., Николаева Е.Н., Ипполитов Е.В. Пародонтопатогенные бактерии как основные факторы возникновения и развития пародонтита. Журнал микробиологии, эпидемиологии, иммунобиологии. 2017;94(5):101–12. Tsarev V.N., Nikolaeva E.N., Ippolitov E.V. Periodontal pathogenic bacteria as the main factors in the occurrence and development of periodontitis. Journal of Microbiology, Epidemiology and Immunobiology. 2017;94(5):101–12. DOI: https://doi.org/10.36233/0372-9311-2017-5-101-112 EDN: https://elibrary.ru/ctbcar
- Howard K.C., Gonzalez O.A., Garneau-Tsodikova S. Porphyromonas gingivalis: where do we stand in our battle against this oral pathogen? RSC Med. Chem. 2021;12(5):666–704. DOI: https://doi.org/10.1039/d0md00424c
- Zhou M., Rong R., Munro D., et al. Investigation of the effect of type 2 diabetes mellitus on subgingival plaque microbiota by high-throughput 16S rDNA pyrosequencing. PLoS One. 2013;8(4):e61516. DOI: https://doi.org/10.1371/journal.pone.0061516
- Dewhirst F.E., Chen T., Izard J., et al. The human oral microbiome. J. Bacteriol. 2010;192(19):5002–17. DOI: https://doi.org/10.1128/jb.00542-10
- Griffen A.L., Beall C.J., Campbell J.H., et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6(6):1176–85. DOI: https://doi.org/10.1038/ismej.2011.191
- Jalava J., Eerola E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int. J. Syst. Bacteriol. 1999;49(Pt. 4):1375–9. DOI: https://doi.org/10.1099/00207713-49-4-1375
- Янушевич О.О., Царев В.Н., Николаева Е.Н. и др. Первый отечественный опыт выявления ассоциации анаэробных бактерий Filifactor alocis и Porphyromonas gingivalis молекулярно-биологическими методами при заболеваниях пародонта и коморбидной патологии (сравнительное исследование). Вестник Российской академии медицинских наук. 2022;77(6):437–46. Yanushevich O.O., Tsarev V.N., Nikolaeva E.N., et al. The first domestic experience of detecting the association of anaerobic bacteria Filifactor alocis and Porphyromonas gingivalis by molecular biological methods in periodontal diseases and comorbid pathology (comparative research). Annals of the Russian academy of medical sciences. 2022;77(6):437–46. DOI: https://doi.org/10.15690/vramn2262 EDN: https://elibrary.ru/pzajok
- Uematsu H., Sato N., Hossain M.Z., et al. Degradation of arginine and other amino acids by butyrate-producing asaccharolytic anaerobic G am-positive rods in periodontal pockets. Arch. Oral Biol. 2003;48(6):423–9. DOI: https://doi.org/10.1016/s0003-9969(03)00031-1
- Aruni A.W., Roy F., Fletcher H.M. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect. Immun. 2011;79(10):3872–86. DOI: https://doi.org/10.1128/iai.05631-11
- Moffatt C.E., Whitmore S.E., Griffen A.L., et al. Filifactor alocis interactions with gingival epithelial cells. Mol. Oral Microbiol. 2011;26(6):365–73. DOI: https://doi.org/10.1111/j.2041-1014.2011.00624.x
- Kim H.Y., Song M.K., Gho Y.S., et al. Extracellular vesicles derived from the periodontal pathogen Filifactor alocis induce systemic bone loss through Toll-like receptor 2. J. Extracell. Vesicles. 2021;10(12):e12157. DOI: https://doi.org/10.1002/jev2.12157
- Aruni A.W., Zhang K., Dou Y., Fletcher H. Proteome analysis of coinfection of epithelial cells with Filifactor alocis and Porphyromonas gingivalis shows modulation of pathogen and host regulatory pathways. Infect. Immun. 2014;82(8):3261–74. DOI: https://doi.org/10.1128/iai.01727-14
- Fine D.H., Markowitz K., Fairlie K., et al. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis and Filifactor alocis are present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J. Clin. Microbiol. 2013;51(9):2850–61. DOI: https://doi.org/10.1128/jcm.00729-13
- Aja E., Mishra A., Dou Y., Fletcher H.M. Role of the Filifactor alocis hypothetical protein FA519 in oxidative stress resistance. Microbiol. Spectr. 2021;9(3):e0121221. DOI: https://doi.org/10.1128/spectrum.01212-21
- Armstrong C.L., Miralda I., Neff A.C., et al. Filifactor alocis promotes neutrophil degranulation and chemotactic activity. Infect. Immun. 2016;84(12):3423–33. DOI: https://doi.org/10.1128/iai.00496-16
- Aruni A.W., Mishra A., Dou Y., et al. Filifactor alocis a new emerging periodontal pathogen. Microbes Infect. 2015;17(7):517–30. DOI: https://doi.org/10.1016/j.micinf.2015.03.011
- Graetz C., Mann L., Krois J., et al. Comparison of periodontitis patients’ classification in the 2018 versus 1999 classification. J. Clin. Periodontol. 2019;46(9):908–17. DOI: https://doi.org/10.1111/jcpe.13157
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2012;35(Suppl. 1):S64–71. DOI: https://doi.org/10.2337/dc12-s064
- Callahan B.J., McMurdie P.J., Rosen M.J., et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581–3. DOI: https://doi.org/10.1038/nmeth.3869
- Quast C., Pruesse E., Yilmaz P., et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database):D590–6. DOI: https://doi.org/10.1093/nar/gks1219
- Tang Z.Z., Chen G., Alekseyenko A.V. PERMANOVA-S: association test for microbial community composition that accommodates confounders and multiple distances. Bioinformatics. 2016;32(17):2618–25. DOI: https://doi.org/10.1093/bioinformatics/btw311
- Langille M.G., Zaneveld J., Caporaso J.G., et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013;31(9):814–21. DOI: https://doi.org/10.1038/nbt.2676
- Parks D.H., Tyson G.W., Hugtnholtz P., Beiko R.G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30(21):3123–4. DOI: https://doi.org/10.1093/bioinformatics/btu494
- Saeb A.T.M., Al-Rubeaan K.A., Aldosary K., et al. Relative reduction of biological and phylogenetic diversity of the oral microbiota of diabetes and pre-diabetes patients. Microb. Pathog. 2019;128:215–29. DOI: https://doi.org/10.1016/j.micpath.2019.01.009
- Casarin R.C., Barbagallo A., Meulman T., et al. Subgingival biodiversity in subjects with uncontrolled type-2 diabetes and chronic periodontitis. J. Periodontal Res. 2013;48(1):30–6. DOI: https://doi.org/10.1111/j.1600-0765.2012.01498.x
- Tam J., Hoffmann T., Fischer S., et al. Obesity alters composition and diversity of the oral microbiota in patients with type 2 diabetes mellitus independently of glycemic control. PLoS One. 2018;13(10):e0204724. DOI: https://doi.org/10.1371/journal.pone.0204724
- Tang B., Yan C., Shen X., Li Y. The bidirectional biological interplay between microbiome and viruses in periodontitis and type 2 diabetes mellitus. Front. Immunol. 2022;13:885029. DOI: https://doi.org/10.3389/fimmu.2022.885029
- Ganesan S.M., Joshi V., Fellows M., et al. A tale of two risks: smoking, diabetes and the subgingival microbiome. ISME J. 2017;11(9):2075–89. DOI: https://doi.org/10.1038/ismej.2017.73
- Uematsu H., Sato N., Hossain M.Z., et al. Degradation of arginine and other amino acids by butyrate-producing asaccharolytic anaerobic Gram-positive rods in periodontal pockets. Arch. Oral Biol. 2003;48(6):423–9. DOI: https://doi.org/10.1016/s0003-9969(03)00031-1
- Cueno M.E., Ochiai K. Gingival periodontal disease (PD) level-butyric acid affects the systemic blood and brain organ: Insights into the systemic inflammation of periodontal disease. Front. Immunol. 2018;9:1158. DOI: https://doi.org/10.3389/fimmu.2018.01158
- Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014;35(1):3–11. DOI: https://doi.org/10.1016/j.it.2013.09.001
- Imai K., Ogata Y. How does Epstein-Barr virus contribute to chronic periodontitis? Int. J. Mol. Sci. 2020;21(6):1940. DOI: https://doi.org/10.3390/ijms21061940
- Li Y., He J., He Z., et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 2014;8(9):1879–91. DOI: https://doi.org/10.1038/ismej.2014.28
- Fang Y., Xu J., Li Z., et al. Design and synthesis of novel pyrimido[5,4-d]pyrimidine derivatives as GPR119 agonist for treatment of type 2 diabetes. Bioorg. Med. Chem. 2018;26(14):4080–7. DOI: https://doi.org/10.1016/j.bmc.2018.06.035
Supplementary files