Genomic features of resistant Klebsiella pneumonia, isolated from the bloodstream and cerebrospinal fluid of pediatric hospital patients
- Authors: Voronina O.L.1, Kunda M.S.1, Ryzhova N.N.1, Aksenova E.I.1, Sadeeva Z.Z.2, Novikova I.E.2, Lazareva A.V.2, Karaseva O.V.2,3, Fisenko А.P.2, Gintsburg A.L.1
-
Affiliations:
- N.F. Gamaleya National Research Center for Epidemiology and Microbiology
- National Medical Research Center for Children's Health
- Clinical and Research Institute of Emergency Pediatric Surgery and Trauma
- Issue: Vol 100, No 6 (2023)
- Pages: 399-409
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/16594
- DOI: https://doi.org/10.36233/0372-9311-430
- EDN: https://elibrary.ru/ylxbdz
- ID: 16594
Cite item
Abstract
Introduction. Carbapenemase-producing Klebsiella pneumoniae (CP-Kp), which are international high-risk clones, have become a problem of utmost importance. CP-Kps, adapting to the hospital environment, evolve into convergent pathotypes. Such variants combine traits of two genetic lineages: multidrug resistant (MDR) and hypervirulent. The pathotypes, along with MDR K. pneumoniae, pose an exceptional threat to young patients during systemic infection.
The objective of this study is the detailed molecular genetic analysis of MDR isolates of K. pneumoniae detected during the monitoring of resistant Gram-negative bacteria at the National Medical Research Center for Children’s Health in 2014–2021.
Materials and methods. Whole-genome sequencing with a subsequent bioinformatics analysis of eight MDR isolates from the bloodstream and cerebrospinal fluid.
Results. MDR isolates belonged to 4 sublineages (SL): SL307, SL395, SL29 and SL1198. In the genomes of 6 pangrug-resistant (PDR) isolates, genes associated with resistance to all categories of antibiotics recommended for Enterobacteriaceae therapy were identified. Plasmids were present in all genomes. In 6 isolates, plasmids contained heavy metal ion resistance operons in addition to antibiotic resistance genes. Prophages within the plasmids were also involved in the transfer of resistance genes. The ST395 isolate from the cerebrospinal fluid belonged to the convergent pathotype in terms of resistance and virulence. Comparison of genomes within SLs revealed recombination events in the K- and O-locus regions and the Yersiniabactin operon.
Conclusion. Thus, in a sample of resistant K. pneumoniae isolated from bloodstream and cerebrospinal fluid, 6 PDR isolates were detected, one of which belongs to the convergent pathotype ST395.
Full Text
Introduction
Klebsiella pneumoniae occupies the leading position among the Enterobacteriaceae causing nosocomial infections in Russia [1]. A concerning problem is the growing proportion of carbapenem-resistant (carbapenemase-producing, CP-Kp) K. pneumoniae and the identification of isolates with carbapenemase genes of different classes (A, B, D) simultaneously present in their genomes [1]. According to the data of 2020, CP-Kp in Russia belonged to clonal groups (CG) CG395, CG11, CG147, and CG307 [1], which are classified as international high-risk clones. CP-Kp ST395 prevail in bloodstream infections in patients with hematologic malignancies (37.7%) [2], neurosurgery ICU patients (27.1%) [3], causing severe systemic infections [4]. During the years 2013 to 2018, the AMRmap — national antibiotic resistance monitoring system (https://AMRmap.ru) — identified 108 CP-Kp ST395 isolates included in an international genomic study that showed the emergence of a convergent pathotype in CG395, combining markers of resistance and hypervirulence [5]. Convergence of traits of two genetic lineages of K. pneumonia, being multidrug resistant (MDR) and hypervirulent (hvKp), was first reported by Gu et al. for isolate ST11 [6]. Starkova et al. studied convergent (CP- hvKp) K. pneumonia isolates ST15, ST147, ST395, ST874, identified in clinics of St. Petersburg, and showed the presence of hybrid plasmids carrying virulence genes and NDM β-lactamase within them [7]. Along with plasmids, other mobile genetic elements such as phages, integrons and transposons contribute to the evolution of bacterial genomes [8]. Conjugative transposons (integrative conjugative elements, ICE) play a specific role in horizontal gene transfer in K. pneumoniae [9]. Such a high rate of adaptation and evolution of K. pneumoniae in hospital settings poses a particular threat to young patients in need of surgical intervention. Bloodstream infections in such patients lead to unfavorable clinical prognosis [10]. The objective of our study was to analyze MDR K. pneumoniae isolated from the bloodstream and cerebrospinal fluid of patients at the National Medical Research Center (NMRC) for Children’s Health in 2014–2021.
Materials and methods
Materials
8 cultures of K. pneumoniae (Table 1) isolated from the bloodstream and cerebrospinal fluid at the NMRC for Children's Health and the Research Institute of Emergency Pediatric Surgery and Traumatology during monitoring of drug resistance of nosocomial isolates of K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa (KAP) in 2014-2021. The study was carried out with the voluntary informed consent of patients and their legal representatives, the study protocol was approved by the Local Ethics Committee of the NMRC for Children's Health (protocol No. 5a dated 02.06.2022). The cultures were characterized by phenotypic multidrug resistance (MDR), extensive drug resistance (XDR) or pandrug Resistance (PDR) in relation to the antibiotic categories recommended by EUCAST for Enterobacteriaceae [11, 12], and according to the EUCAST criteria [11].
Table 1. Characteristics of antibiotic-resistant K. pneumoniae isolates
Patient | Isolate | Data of isolation | Accession number | Genotype | Sublineage | Clonal group (CG) | wzi allele | Associated KL type | O locus | O type | Virulence score | Resistance scores |
P9 | SCCH70:Kpn76815 | 19.06.2017 | JAUPHS000000000 | ST307 | SL307 | CG307 | – | KL102* | O1/O2v2 | O2afg | 1 | 3 |
P11 | SCCH72:Kpn881723 | 20.09.2018 | JAUPHU000000000 | ST2975 | SL307 | CG307 | 173 | KL102 | OL102 | OL102 | 1 | 3 |
P8 | SCCH69:Kpn69138 | 13.04.2016 | JAUOCC000000000 | ST395 | SL395 | CG395 | 2 | KL2 | O1/O2v2 | O2afg | 4 | 3 |
P18 | SCCH87:Kpn2182174 | 05.11.2021 | CP122350-CP122355 | ST395 | SL395 | CG395 | 2 | KL2 | O1/O2v1 | O1 | 1 | 3 |
P17 | SCCH84:Kpn2082401 | 12.09.2020 | JAUTIO000000000 | ST866 | SL1198 | CG1198 | 478 | KL46 | O3b | O3b | 1 | 3 |
P17 | SCCH86:Kpn207262 | 02.10.2020 | JAUTIQ000000000 | ST866 | SL1198 | CG1198 | 478 | KL46 | O3b | O3b | 1 | 3 |
P10 | SCCH71:Kpn863165 | 17.12.2018 | JAUPHT000000000 | ST985 | SL29 | CG985 | 39 | KL39 | O1/O2v2 | O1 | 1 | 3 |
P12 | SCCH73:Kpn96857 | 11.03.2019 | JAUPHV000000000 | ST29 | SL29 | CG29 | 19 | KL19 | O1/O2v2 | O1 | 1 | 3 |
Methods
Cultivation of K. pneumoniae was carried out on blood and Uri-select agar (BioRad) at 37ºС for 24-48 hours. MALDI-TOF mass spectrometer (Bruker Daltonics) was used for identification.
Minimum inhibitory concentrations (MICs) were determined by microdilution for the following categories of antimicrobials:
- Aminoglycosides: gentamicin, tobramycin, amikacin, netilmicin;
- Carbapenems: imipenem, meropenem;
- Extended-spectrum (3rd and 4th generation) cephalosporins: cephalosporin, cefepime;
- Fluoroquinolones: ciprofloxacin, levofloxacin;
- Folate metabolism inhibitors: trimethoprim/sulfamethoxazole;
- Monobactams: aztreonam;
- Phosphonic acids: fosfomycin;
- Polymyxins: colistin (polymyxin E), polymyxin B;
- Penicillins with beta-lactamase inhibitors: ticarcillin/clavulanate, piperacillin/tazobactam.
To identify resistance mechanisms and analyze virulence factors, 8 isolates were examined by whole-genome sequencing.
DNA was extracted from isolates using a method described elsewhere [13], supplemented by purification from polysaccharides using CTAB (cetyltrimethylammonium bromide).
DNA libraries were prepared according to the protocol of Nextera DNA Flex Library Prep (Illumina) and sequenced on a NextSeq 500/550 instrument (Illumina) using a "Mid Output 300 cycles" cartridge.
CLC Genomic Workbench v.21.0.1 (Qiagen) and SPAdes v3.13.01 were used for genome assembly. CGView Server was used to visualize the results of replicon assembly and genome comparisons2 [14]. Annotation was performed using Rapid Annotations Subsystems Technology [15] and NCBI Prokaryotic Genome Annotation Pipeline [16]. PHASTER was used to search for prophage areas3 [17]. Results of full-genome sequencing were deposited in GenBank in bioproject PRJNA561493 under the numbers shown in Table 1.
Genomic data were analyzed using the resources of the platform Pathogenwatch v. 21.0.04, which allows interaction with Kleborate v. 2.2.05 [18], a resource specifically designed for K. pneumoniae complex research. Kleborate v. 2.2.0 was used to determine genotype in the context of MLST (MultiLocus Sequence Typing) genes, virulence factors: yersiniabactin (ybt), aerobactin (iuc), colibactin (clb), salmohelin (iro), hypermucoidity (rmpA, rmpA2), as well as antimicrobial resistance determinants. With reference to the Kaptive site6 [19] alleles of the wzi gene were determined, and K loci (capsule) and O loci (lipopolysaccharide) were typed.
The criteria developed by Kleborate v. 2.2.0 were used to assess the level of virulence and resistance.
For virulence:
- level 5 — ybt, iuc, clb;
- level 4 — ybt, iuc;
- level 3 — iuc;
- level 2 — ybt, clb (or only clb);
- level 1 — ybt;
- level 0 — complete absence.
For resistance:
- level 3 — carbapenemases and colistin resistance;
- level 2 — carbapenemases;
- level 1 — extended-spectrum β-lactamases without carbapenemases;
- level 0 — absence of extended-spectrum β-lactamases [18].
The VFDB7 database was used to analyze the extended spectrum of virulence factors [20]. The web platform BIGSdb-Pasteur8 was accessed to clarify genotypes for ybt, iuc, clb, as well as for the identification of heavy metal ion resistance loci.
A more complete spectrum of resistance genes was determined using the CARD resource9 [21], as well as the BV-BRC10, formed on the basis of PATRIC [22].
PlasmidFinder 2.111 [23] was used to identify incompatibility groups (Inc) of plasmid replicons.
Results
685 cultures were isolated from bloodstream and cerebrospinal fluid during monitoring of drug resistance of nosocomial isolates of K. pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa (KAP) in two multidisciplinary children's hospitals: NMRC for Children's Health and Research Institute of Emergency Pediatric Surgery and Traumatology, in 2014-2021, including 63 (9.2%) isolates of K. pneumoniae. Eight isolates presented in Table 1 were resistant to multiple categories of antibiotics. Isolates from patients P8-P12 and P18 were PDR according to MIC data for all 9 antibiotic categories. Two K. pneumoniae isolated 3 weeks apart from patient P17 were examined. Both isolates were sensitive to fluoroquinolones (to levofloxacin at increased exposure) (Table 2). The first isolate by time of isolation was also sensitive to folate metabolism inhibitors and even showed an MIC against polymyxin B corresponding to a borderline resistance value. Thus, the former isolate can be recognized as MDR and the latter as XDR.
Table 2. Comparison of antibiograms of K. pneumoniae ST866 isolates from patient P17 in three categories of antibiotics
Isolate | MIC, mg/L | |||
fluoroquinolones | folate pathway inhibitors | polymyxins | ||
ciprofloxacin | levofloxacin | trimethoprim/sulfamethoxazole | Polymixin B | |
SCCH84:Kpn2082401 | ≤ 0,25 | 1* | 2 | 2 |
SCCH86:Kpn207262 | 0,5* | 1* | 16/304 | 16 |
Resistance — MIC breakpoints, mg/L | 0,5 | 1 | 4 | 2 |
Note. *The sensitivity at increased exposure; the cells corresponding to the sensitivity of the isolate to the antimicrobial drug are highlighted in grеy.
Out of the 8 isolates, 7 were isolated from the bloodstream and 1 (P8) was isolated from cerebrospinal fluid. Patients, with the exception of P12 (8 years old), were under 1 year of age, predominantly had genetically determined diseases resulting in malformations requiring surgical intervention. P8 had severe combined trauma, P12 had systemic vasculitis accompanied by hemolytic-uremic syndrome. Fatal outcome was recorded in 2 cases (P10 and P12).
The main characteristics of resistant isolates. All 8 isolates belonged to phylogroup Kp1 and were subdivided into 4 sublineages (SL) (Table 1), 2 of which (307 and 395) are among the most represented in nosocomial K. pneumoniae in Russia [1]. Three sublineages included one CG each. SL29 was represented by 2 CGs. Despite the fact that ST985 differs by one locus (gapA) allele profile from ST29 (single locus variant, SLV), it belongs to another clonal group — CG985. It is important to note that in CG307, ST2975 is also the SLV of ST307 at the rpoB locus.
The SL29 isolates had the same O-antigen variant of O1/O2v2. The same variant was characteristic of the ST307 and ST395 P8 isolates. Another ST395 isolate (P18) had an O-antigen of O1/O2v1. The different variants of the O-antigen are determined by the monomer constituting the polysaccharide. Variant 1 contains d-galactan I and variant 2 contains d-galactan III [24]. Thus, even the isolate genomes of the same genotype may differ in the structure of the operon responsible for the O-antigen. P17 isolates have their own variant, O3b, but similar to the O1/O2 variants, it is prevalent in isolates causing human diseases [24]. Isolate ST2975 has a new O-antigen, therefore it is named OL102 after the capsular antigen number.
In contrast, capsular polysaccharides are distinct in SL29 isolates but identical in ST395 isolates. SL307 isolates have a new KL-type, KL102. Furthermore, while the capsular operon in isolate ST2975 is fully represented, the ST307 genome contains only the full-length cpsACP among the genes at the 5'-end of the operon (galF, cpsACP, wzi, wza, wzb, wzc). The galF gene is a pseudogene, and 4 other genes, as well as the following wbaP, are missing. The wbaP gene encodes a glycosyltransferase that initiates capsular polysaccharide synthesis by transferring galactose-1-phosphate to the acceptor udeprenyl-phosphate. In the absence of this enzyme, capsule synthesis is not possible [25]. The wzi and wza genes are responsible for outer membrane channel proteins, while the wzc and wzb genes are regulators of polymerization and transport of capsular polysaccharides to the surface of the bacterial cell.
The fimbriae operons fimA–fimK (type 1) and mrkA–mrkJ (type 3), as well as the type IV pilW pilus gene were present in the genomes of all isolates.
In order to determine the resistance and virulence of the selected isolates based on genomic data, we used the quantitative assessments offered by Kleborate v. 2.2.0 [19] (Table 1). The genomes of all isolates contained the ybt operon, which is located in ICEKp: ICEKp4 (ybt10) in CG307 isolates, ICEKp5 (ybt14) in ST395 P8 and ST29 isolates, ICEKp12 (ybt16) in ST395 P18 and ST985 isolates, and ICEKp15 (ybt18) in ST866 isolates.
Most of the isolates had a virulence level of 1. The ST395 P8 isolate had a level of 4, the highest, because in addition to ybt, the isolate's genome encodes aerobactin (AbST95). Of the additional virulence genes, peg-344 [26], responsible for drug and metabolite transporter permease, is present only in the ST395 P8 genome. The hypermucoidity genes rmpA and rmpA2 were not detected in this group of isolates.
The resistance level is the highest in all isolates (3), because both carbapenemase genes and mutations in genes (PhoP_26Q) providing resistance to colistin (polymyxin E) are present in the genomes [27].
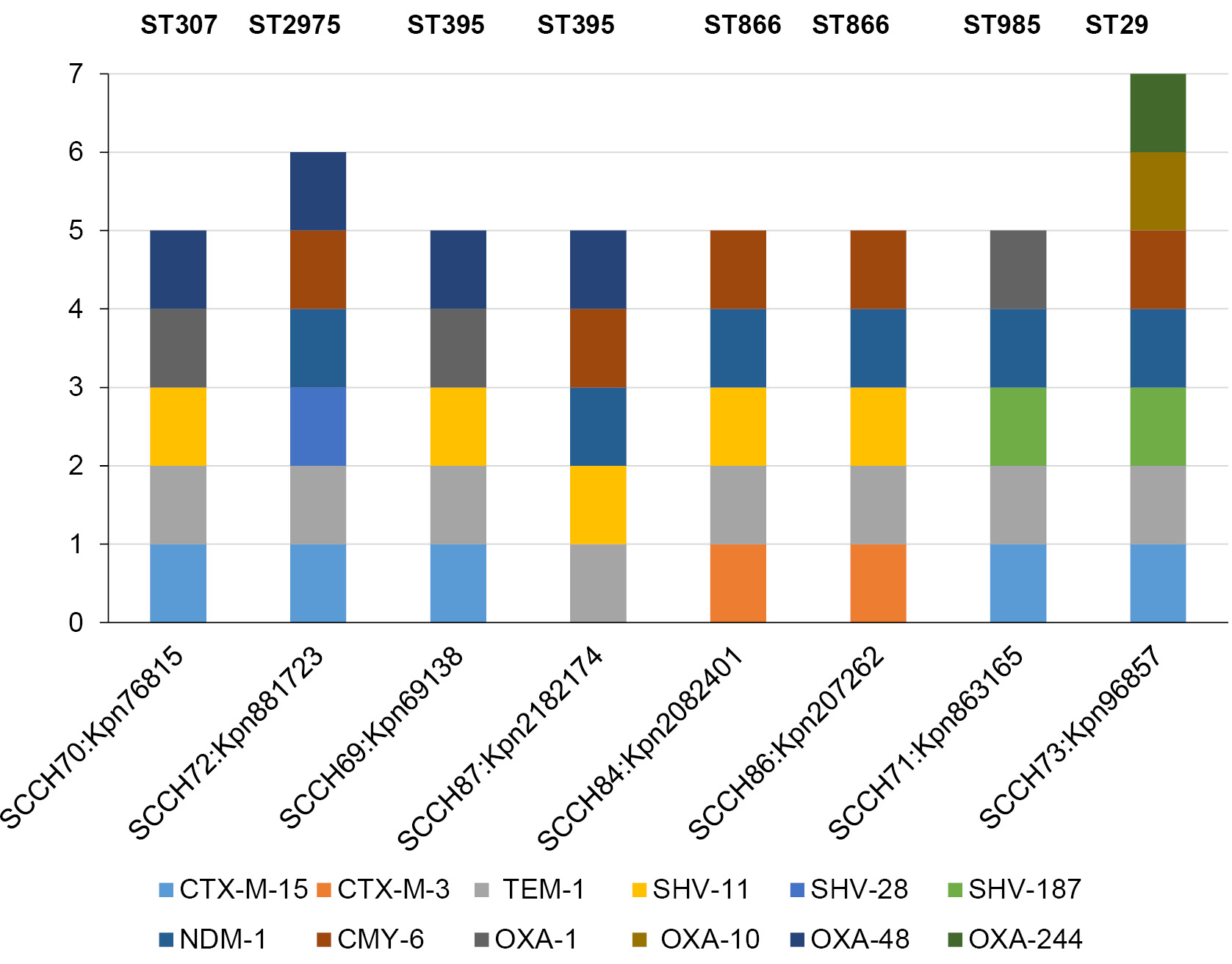
The spectrum of β-lactamase genes in the genomes of isolates is extensive (Fig. 1). All genomes contain at least one carbapenemase: OXA-48 (class D) or NDM-1 (class B, metallo-β-lactamase), and the ST2975 and ST395 P18 genomes contain both β-lactamases. NDM-1 is also classified as an extended-spectrum β-lactamase [28], to which CTX-M-15 and CTX-M-3 (class A) also belong. Only the ST395 P18 isolate did not contain CTX-M lactamase. As far as the other class A β-lactamases is concerned, the TEM and SHV lactamases are present in all genomes. The CMY-6 cephalosporinase (class C) is present in most genomes; it is absent only in the ST307 and ST395 P8 genomes. However, these genomes have an additional class D β-lactamase, OXA-1, which is also present in the ST985 genome. The ST29 genome is distinguished by the presence of two additional class D β-lactamases, OXA-10 and OXA-244. Thus, the isolate genomes contain at least 5 genes of β-lactamases of different classes, while there are 7 genes in the ST29 genome.
Fig. 1. Diversity of beta-lactamase genes in the genomes of the studied isolates.
Resistance genes to other antibiotic categories are shown in Fig. 2. A minimum of 6 additional resistance genes were detected in ST866 isolates. Among them there are no resistance genes to fluoroquinolones and trimethoprim, which confirms the phenotypic data. The maximum number of additional genes in isolate ST395 P8 is 16. Along with isolate ST985 (14 genes), they have at least one resistance-determining gene for each category of antibiotics. It is important to note that only the leading isolates have macrolide resistance genes.
Fig. 2. Diversity of resistance genes to other categories of antibiotics in the genomes of the studied isolates.
Heavy metal ion resistance genes were found in 6 out of 8 genomes (Fig. 3). These are resistance operons to arsenic (arsABCDR), copper (pcoABCDERS), silver (silABCEFGPRS), and tellurium (terABCDEWXYZ). All 4 operons are present in the ST307 genome, 3 each in the ST2975 and ST985 genomes, 2 each in ST866, with only the tellurium resistance operon being present in the ST395 P8 genome.
Fig. 3. Genes of resistance to heavy metal ions in the genomes of the studied isolates. The red dot marks the incomplete arsBCRD operon.
Plasmids of resistant isolates. Typically, heavy metal operons and many resistance genes are present in plasmids [29]. We estimated the number of Inc plasmid groups for the genomic data of each isolate (Fig. 4). The maximum number of Inc was found in isolate ST307 (7) and the minimum in isolates ST985 (1) and ST866 (3). The data confirm the presence of plasmids in the genomes of all isolates.
Fig. 4. Diversity of origins of replication (Inc) plasmids in the studied isolates.
The different spectrum of Inc in isolates of the same ST395 genotype attracts attention. Assembly of plasmid replicons of ST395 P8 and ST395 P18 isolates showed that the ST395 P18 genome contains 5 plasmids, 3 of which are capable of conjugative transfer, and the ST395 P8 genome contains 4 plasmids, one of which is cryptic (2 are capable of conjugative transfer). The plasmidome of the isolates differed in the number and set of resistance and virulence genes (Fig. 5). The plasmidome of ST395 P8 was characterized by the presence of extended spectrum β-lactamase genes, aerobactin, peg-344, tellurium resistance operon, and a greater number of β-lactamase genes. The plasmidome of ST395 P18 had more carbapenemase genes, sulfonamide and aminoglycoside resistance genes. Thus, the plasmidome of ST395 isolates underwent significant changes from 2016 to 2021.
Fig. 5. Resistance and virulence genes in K. pneumoniae ST395 plasmids. Circled are factors that are present only in the plasmids of the ST395 P8 isolate.
Comparison of the prophages of ST395 isolates showed differences in these mobile genetic elements as well. Eight prophages were identified in the ST395 P8 genome, 7 in the ST395 P18 genome; 5 prophages were common to the ST395 genomes. However, ST395 P8 plasmids contained twice as many prophages (10) compared to ST395 P18 plasmids (5). All prophages had homology with 17 different bacterial phages of the Caudoviricetes class. The chromosome prophages each contained 3 holin genes that form pores in the cell membrane [30], and lysin genes that damage peptidoglycan: 4 lysozyme genes, ST395 P8, and 5 lysozyme genes and a membrane-bound lytic transglycosylase D gene, ST395 P18. CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats) systems were absent in both genomes. At the same time, the analysis of prophage plasmids of ST395 isolates confirmed the observation that phages could possibly be carriers of antibiotic resistance genes [31]. The prophages of plasmids of ST395 P8 isolate contained genes of β-lactamases (5), chloramphenicol acetyltransferases (3), aminoglycoside acetyltransferases (2), dihydrofolate reductase, streptomycin adenyltransferase, and tellurium resistance operon. The plasmid prophages of isolate ST395 P18, which contained genes for aminoglycoside acetyltransferases (3), dihydropteroate synthases (2), β-lactamases TEM and OXA-48, chloramphenicol acetyltransferase and 3 SMR (Small multidrug resistance) efflux transporters, were not inferior to them either.
Discussion
The study revealed a great diversity of MDR K. pneumoniae genotypes isolated from cerebrospinal fluid and bloodstream. According to the data of the Pasteur Institute12, ST307 is the most prevalent worldwide (1786 isolates, 21 in Russia), followed by ST395 (380, 188 in Russia) and ST29 (255, 2 in Russia), ST985 (48), ST2975 (29, 12 in Russia), and ST866 (7). Infection with PDR K. pneumoniae ST29 and ST985 resulted in fatal outcomes. The systemic infection was detected in other loci besides the bloodstream in both patients P10, P12 and patients P9, P11, P17, P18. The infection in Patient P8 was detected only in the cerebrospinal fluid and was caused by the most virulent ST395 isolate (level 4). In terms of resistance (PDR) and virulence, the ST395
P8 isolate matched the characteristics of the convergent ST395 pathotype [5]. The study of the isolate plasmids confirms the transfer of resistance factors (including to heavy metal ions) and virulence factors by these mobile genetic elements and prophages within the plasmids. Comparison of the ST395 P8 and ST395 P18 genomes reveals evidence of recombination in hot spots characterized by E.R. Shaidullina et al. [5]. The O-antigen, ICEKp and prophage regions differ among isolates. Isolate ST395 P18 has ICEKp12 (ybt16) prevalent in CG305 and in subclade B1 (KL2KL30) [5], with the K-locus characterization of the isolate corresponding to that of the subclade. In the ST395 P8 isolate, ICEKp5 (ybt14) is rare [5] and belongs to subclade A1 (KL108). However, the ST395 P8 isolate contains a KL2 K-locus, which may be further evidence of recombination events.
In CG307 isolates, differences in the set of resistance genes are associated with a different plasmid profile. In the chromosome, the isolates contain the same ICEKp4 (ybt10) regions, but significant differences in the main recombination hot spot, the K- and O-loci.
In SL29, the isolates are similar only in the O-locus, have different K-loci, ICEKp, plasmid profile and resistance gene spectrum. It is important to note that the single plasmid of isolate ST985 (IncFIB(K)) contains 3 heavy metal ion resistance operons and a large list of genes that define together with genes of the chromosome pandrug-resistance of the isolate.
ST866 isolates, while being the least resistant, are nevertheless MDR and XDR, carrying plasmids containing 2 operons of resistance to heavy metal ions.
Returning to the actual problem of resistance of K. pneumoniae to carbapenems, it is necessary to note the presence of 2 isolates containing 2 carbapenemase genes each: OXA-48 and NDM-1, in the sample. The isolates belong to international high-risk clones CG395 and CG307.
Thus, in the sample of resistant isolates of K. pneumoniae isolated from bloodstream and cerebrospinal fluid, 6 PDR isolates were detected, one of which belongs to the convergent pathotype ST395.
1 St. Petersburg genome assembler, Russia,
URL: http://cab.spbu.ru/software/spades/
2 CGView Server.
URL: http://stothard.afns.ualberta.ca/cgview_server/
3 PHAge Search Tool Enhanced Release. URL: https://phaster.ca/
4 Pathogenwatch v. 21.0.0.
5 Kleborate v. 2.2.0.
URL: https://github.com/klebgenomics/Kleborate/wiki
6 Kaptive. URL: https://kaptive-web.erc.monash.edu/
7 Virulence factor database. URL: http://www.mgc.ac.cn/VFs/
8 Institut Pasteur Klebsiella pneumoniae species complex.
URL: https://bigsdb.pasteur.fr/klebsiella/
9 Comprehensive Antibiotic Resistance Database.
URL: https://card.mcmaster.ca/
10 Bacterial and Viral Bioinformatics Resource Center.
11 PlasmidFinder 2.1.
URL: https://cge.food.dtu.dk/services/PlasmidFinder/
12 Institut Pasteur. URL: bigsdb.pasteur.fr/klebsiella
About the authors
Olga L. Voronina
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: olv550@gmail.com
ORCID iD: 0000-0001-7206-3594
Cand. Sci. (Biol.), Assistant Professor, Head, Laboratory of genome analysis
Russian Federation, MoscowMarina S. Kunda
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Author for correspondence.
Email: markunda99@gmail.com
ORCID iD: 0000-0003-1945-0397
Cand. Sci. (Biol.), senior researcher, Laboratory of genome analysis
Russian Federation, MoscowNatalia N. Ryzhova
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: rynatalia@yandex.ru
ORCID iD: 0000-0001-5361-870X
Cand. Sci. (Biol.), senior researcher, Laboratory of genome analysis
Russian Federation, MoscowEkaterina I. Aksenova
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: aksenova16@yandex.ru
ORCID iD: 0000-0003-2704-6730
Cand. Sci. (Biol.), senior researcher, Laboratory of genome analysis
Russian Federation, MoscowZulfirya Z. Sadeeva
National Medical Research Center for Children's Health
Email: zulfiryasadeeva@yandex.ru
ORCID iD: 0000-0002-4587-0902
junior researcher, Laboratory of molecular microbiology
Russian Federation, MoscowIrina E. Novikova
National Medical Research Center for Children's Health
Email: novikovayudina@outlook.com
ORCID iD: 0000-0003-4234-0209
junior researcher, Laboratory of molecular microbiology
MoscowAnna V. Lazareva
National Medical Research Center for Children's Health
Email: lasarevaav@nczd.ru
ORCID iD: 0000-0003-3896-2590
D. Sci. (Med.), chief researcher, Laboratory of molecular microbiology, Head, Laboratory of microbiology
Russian Federation, MoscowOlga V. Karaseva
National Medical Research Center for Children's Health; Clinical and Research Institute of Emergency Pediatric Surgery and Trauma
Email: karaseva.o@list.ru
ORCID iD: 0000-0001-9418-4418
D. Sci. (Med.), Head, Department of emergency surgery and childhood injuries, National Medical Research Center for Children's Health, Moscow, Russia; Deputy director for scientific work, Head, Department of combined trauma, anesthesiology-resuscitation, Clinical and Research Institute of Emergency Pediatric Surgery and Trauma
Russian Federation, Moscow; MoscowАndrey P. Fisenko
National Medical Research Center for Children's Health
Email: olv550@gmail.com
ORCID iD: 0000-0001-8586-7946
D. Sci. (Med.), Professor, Director
Russian Federation, MoscowAlexander L. Gintsburg
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: gintsburg@gamaleya.org
ORCID iD: 0000-0003-1769-5059
D. Sci (Biol.), Professor, Academician of the Russian Academy of Sciences, Director
Russian Federation, MoscowReferences
- Белобородов В.Б., Голощапов О.В., Гусаров В.Г. и др. Методические рекомендации Российской некоммерческой общественной организации «Ассоциация анестезиологов-реаниматологов», Межрегиональной общественной организации «Альянс клинических химиотерапевтов и микробиологов», Межрегиональной ассоциации по клинической микробиологии и антимикробной химиотерапии (МАКМАХ), общественной организации «Российский Сепсис Форум» «Диагностика и антимикробная терапия инфекций, вызванных полирезистентными штаммами микроорганизмов» (обновление 2022 г.). Вестник анестезиологии и реаниматологии. 2022;19(2):84–114. Beloborodov V.B., Goloschapov O.V., Gusarov V.G., et al. Guidelines of the Association of anesthesiologists-intensivists, the Interregional non-governmental organization Alliance of clinical chemotherapists and microbiologists, the Interregional Association for Clinical Microbiology and Antimicrobial Chemotherapy (IACMAC), and NGO Russian sepsis forum diagnostics and antimicrobial therapy of the infections caused by multiresistant microorganisms (update 2022). Messenger of Anesthesiology and Resuscitation. 2022;19(2):84–114. DOI: https://doi.org/10.21292/2078-5658-2022-19-2-84-114 EDN: https://elibrary.ru/vjuogq
- Khrulnova S., Fedorova A., Frolova I., et al. Distribution of virulence genes and capsule types in Klebsiella pneumoniae among bloodstream isolates from patients with hematological malignancies. Diagn. Microbiol. Infect. Dis. 2022;104(1):115744. DOI: https://doi.org/10.1016/j.diagmicrobio.2022.115744
- Fursova A.D., Fursov M.V., Astashkin E.I., et al. Early response of antimicrobial resistance and virulence genes expression in classical, hypervirulent, and hybrid hvKp-MDR Klebsiella pneumoniae on antimicrobial stress. Antibiotics (Basel). 2021;11(1):7. DOI: https://doi.org/10.3390/antibiotics11010007
- Fursova N.K., Astashkin E.I., Ershova O.N., et al. Multidrug-resistant Klebsiella pneumoniae causing severe infections in the Neuro-ICU. Antibiotics (Basel). 2021;10(8):979. DOI: https://doi.org/10.3390/antibiotics10080979
- Shaidullina E.R., Schwabe M., Rohde T., et al. Genomic analysis of the international high-risk clonal lineage Klebsiella pneumoniae sequence type 395. Genome Med. 2023;15(1):9. DOI: https://doi.org/10.1186/s13073-023-01159-6
- Gu D., Dong N., Zheng Z., et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 2018;18(1):37–46. DOI: https://doi.org/10.1016/S1473-3099(17)30489-9
- Starkova P., Lazareva I., Avdeeva A., et al. Emergence of hybrid resistance and virulence plasmids harboring New Delhi metallo-β-lactamase in Klebsiella pneumoniae in Russia. Antibiotics (Basel). 2021;10(6):691. DOI: https://doi.org/10.3390/antibiotics10060691
- Khedkar S., Smyshlyaev G., Letunic I., et al. Landscape of mobile genetic elements and their antibiotic resistance cargo in prokaryotic genomes. Nucleic Acids Res. 2022;50(6):3155–68. DOI: https://doi.org/10.1093/nar/gkac163
- Johnson C.M., Grossman A.D. Integrative and Conjugative Elements (ICEs): What they do and how they work. Annu. Rev. Genet. 2015;49:577–601. DOI: https://doi.org/10.1146/annurev-genet-112414-055018
- Shapaka J.T., Muloiwa R., Buys H. Association of full blood count findings with risk of mortality in children with Klebsiella pneumoniae bloodstream infection at a South African children's hospital. BMC Pediatr. 2023;23(1):302. DOI: https://doi.org/10.1186/s12887-023-04104-z
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 13.1; 2023. URL: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_13.1_Breakpoint_Tables.pdf
- Magiorakos A.P., Srinivasan A., Carey R.B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18(3):268–81. DOI: https://doi.org/10.1111/j.1469-0691.2011.03570.x
- Wilson K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001;Chapter 2:Unit2.4. DOI: https://doi.org/10.1002/0471142727.mb0204s56
- Grant J.R., Stothard P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36(Web Server issue):W181–4. DOI: https://doi.org/10.1093/nar/gkn179
- Brettin T., Davis J.J., Disz T., et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. DOI: https://doi.org/10.1038/srep08365
- Li W., O'Neill K.R., Haft D.H., et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49(D1):D1020–8. DOI: https://doi.org/10.1093/nar/gkaa1105
- Arndt D., Grant J.R., Marcu A., et al. PHASTER: а better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–21. DOI: https://doi.org/10.1093/nar/gkw387
- Lam M.M.C., Wick R.R., Watts S.C., et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021;12(1):4188. DOI: https://doi.org/10.1038/s41467-021-24448-3
- Lam M.M.C., Wick R.R., Judd L.M., et al. Kaptive 2.0: updated capsule and lipopolysaccharide locus typing for the Klebsiella pneumoniae species complex. Microb. Genom. 2022;8(3):000800. DOI: https://doi.org/10.1099/mgen.0.000800
- Chen L., Yang J., Yu J., et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33(Issue suppl. 1):D325–8. DOI: https://doi.org/10.1093/nar/gki008
- Alcock B.P., Huynh W., Chalil R., et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023;51(D1):D690–9. DOI: https://doi.org/10.1093/nar/gkac920
- Olson R.D., Assaf R., Brettin T., et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023;51(D1):D678–89. DOI: https://doi.org/10.1093/nar/gkac1003
- Carattoli A., Zankari E., García-Fernández A., et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58(7):3895–903. DOI: https://doi.org/10.1128/AAC.02412-14
- Follador R., Heinz E., Wyres K.L., et al. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2016;2(8):e000073. DOI: https://doi.org/10.1099/mgen.0.000073
- Pan Y.J., Lin T.L., Chen C.T., et al. Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 2015;5:15573. DOI: https://doi.org/10.1038/srep15573
- Liu C., Du P., Xiao N., et al. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence. 2020;11(1):1215–24. DOI: https://doi.org/10.1080/21505594.2020.1809322
- Olaitan A.O., Morand S., Rolain J.M. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 2014;5:643. DOI: https://doi.org/10.3389/fmicb.2014.00643
- Palzkill T. Metallo-β-lactamase structure and function. Ann. N.Y. Acad. Sci. 2013;1277:91–104. DOI: https://doi.org/10.1111/j.1749-6632.2012.06796.x
- Wu F., Ying Y., Yin M., et al. Molecular characterization of a multidrug-resistant Klebsiella pneumoniae strain R46 isolated from a rabbit. Int. J. Genomics. 2019;2019:5459190. DOI: https://doi.org/10.1155/2019/5459190
- Fernandes S., São-José C. More than a hole: the holin lethal function may be required to fully sensitize bacteria to the lytic action of canonical endolysins. Mol. Microbiol. 2016;102(1):92–106. DOI: https://doi.org/10.1111/mmi.13448
- Balcazar J.L. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014;10(7):e1004219. DOI: https://doi.org/10.1371/journal.ppat.1004219
Supplementary files