Том 97, № 6 (2020)
- Год: 2020
- Выпуск опубликован: 20.01.2021
- Статей: 16
- URL: https://microbiol.crie.ru/jour/issue/view/41
Весь выпуск
ОРИГИНАЛЬНЫЕ ИССЛЕДОВАНИЯ
Генетическая вариабельность SARS-CoV-2 в биологических образцах от пациентов г. Москвы
Аннотация
Сосуществование субпопуляций SARS-CoV-2 с различными вариантами генома внутри организма одного пациента — один из обсуждаемых в настоящее время феноменов. В данной работе мы провели высокопроизводительное секвенирование и сборку полных геномов вирусов из образцов, которые представляли собой мазки или аутопсийный материал от пациентов с диагнозом СOVID-19, предварительно подтвержденным методом полимеразной цепной реакции в реальном времени (Ct = 10,4–19,8). Подготовку образцов к секвенированию проводили с помощью протокола SCV-2000bp. Полученные данные проверяли на присутствие в образце более чем одного генетического варианта SARS-CoV-2. Варианты нуклеотидных замен, покрытие для каждого варианта, а также координаты вариабельной позиции в референсном геноме определяли с помощью инструментов программы «CLC Genomics Workbench». При поиске вариабельных нуклеотидных позиций исходили из предположения, что в образце имеются 2 генетических варианта (не более), для вероятности определяемого варианта использовали пороговое значение ≥ 90%. Также игнорировали варианты, которые были представлены менее чем 20% прочтений от общего покрытия. Полученные результаты показали, что в 5 образцах имеется вариабельность, т.е. содержится несколько генетических вариантов SARS-CoV-2. В 4 образцах оба найденных варианта геномов различались лишь в одной нуклеотидной позиции. В пятом образце были найдены более существенные различия: сразу 3 вариабельных позиции и одна делеция длиной в 3 нуклеотида. Наше исследование показывает возможность сосуществования различных генетических вариантов SARS-CoV-2 в организме пациента.
 511-517
511-517


Характеристика серопревалентности к SARS-СoV-2 среди населения Республики Татарстан на фоне COVID-19
Аннотация
В конце 2019 г. появились сообщения о вспышке инфекции, вызванной новым штаммом β-коронавируса SARS-CoV-2, заболевание ВОЗ определила как coronavirus disease 2019 (COVID-19). В Республике Татарстране первый случай COVID-19 был диагностирован 16.03.2020 г., это был завозной случай из Франции. Период нарастания заболеваемости продолжался с 12-й по 19-ю неделю, когда был зарегистрирован самый высокий показатель, составивший 16,7 на 100 тыс. населения. В дальнейшем отмечалось статистически значимое снижение заболеваемости. Исследование серопревалентности было проведено на 27-й неделе (8-я неделя снижения заболеваемости)
Целью проведенного сероэпидемиологического исследования было определение уровня и структуры популяционного иммунитета к вирусу SARS-CoV-2 среди населения Республики Татарстан в период распространения COVID-19.
Материалы и методы. Отбор волонтеров для исследования проводили методом анкетирования и рандомизации путем случайной выборки. Критерием исключения была активная инфекция COVID-19 в момент анкетирования. На наличие специфических антител к SARS-CoV-2 обследовано 2946 человек. Возраст обследованных добровольцев варьировал от 1 года до 70 лет и старше.
Результаты. Результаты исследования показали, что в Республике Татарстан в период заболеваемости COVID-19 наблюдалась умеренная серопревалентность к SARS-CoV-2, составившая 31,3%, на фоне высокой частоты (94,5%) бессимптомной инфекции у серопозитивных лиц, не имевших в анамнезе перенесенного заболевания COVID-19, положительного результата ПЦР и симптомов ОРВИ в день обследования. Максимальные показатели коллективного иммунитета установлены у детей 7–13 лет (42,0%), детей 14–17 лет (40,3%) при одновременном снижении серопревалентности у лиц в возрасте 70 лет и старше (24,0%). В разных регионах Республики Татарстан наблюдалось широкое варьирование показателей серопозитивности от минимального в Заинском районе (8,6%) до максимального в Арском районе (74,3%). В 21 районе из 38 обследованных результаты были нерепрезентативны из-за малого числа наблюдений. У реконвалесцентов COVID-19 антитела вырабатываются в 83,3% случаев. У лиц с позитивным результатом ПЦР-анализа, проведенного ранее, антитела выявлялись в 100% случаев. Среди волонтеров, имевших контакты с больными COVID-19, доля серопревалентных 37%.
Вывод. Динамику серопревалентности среди населения Республики Татарстан можно квалифицировать как позитивную. Полученные результаты могут быть использованы для разработки прогноза развития эпидемиологической ситуации, а также планирования мероприятий по специфической и неспецифической профилактике COVID-19.
 518-528
518-528


Чувствительность биопленок вакцинных и свежевыделенных штаммов Bordetella pertussis к антибиотикам
Аннотация
Цель. Изучение чувствительности биопленок вакцинных и свежевыделенных штаммов Bordetella pertussis к антибиотикам.
Материалы и методы. Использованы вакцинные и свежевыделенные штаммы B. pertussis. В качестве инокулята для получения биопленок использовали культуры штаммов, выращенные на плотной питательной среде. Интенсивность образования биопленок в круглодонных полистироловых 96-луночных планшетах оценивали окрашиванием 0,1% раствором генцианвиолета. В опытах использовали антибиотики следующих групп: пенициллины (ампициллин), цефалоспорины (цефтриаксон), аминогликозиды (гентамицин), макролиды (эритромицин).
Результаты. Наиболее высокой устойчивостью к антибиотикам отличались вакцинный штамм № 305 и свежевыделенный штамм № 211, проявлявшие чувствительность только к эритромицину. Вакцинный штамм № 703 был чувствителен к гентамицину и ампициллину и проявлял резистентность к эритромицину и цефтриаксону. Вакцинный штамм № 475 был чувствителен ко всем испытанным антибиотикам. Штамм Tohama 1 был резистентен к ампициллину и чувствителен к остальным антибиотикам. Свежевыделенные штаммы № 178 и № 162 были устойчивы к цефтриаксону и чувствительны к гентамицину, эритромицину и ампициллину. Минимальные подавляющие концентрации использованных антибиотиков составляли 0,2–5 мкг/мл.
Заключение. Приведенные данные свидетельствуют о гетерогенности вакцинных и свежевыделенных штаммов B. pertussis по чувствительности к антибиотикам. Наибольшую активность проявлял эритромицин, подавлявший рост биопленок 6 из 7 штаммов. Наименее эффективным был цефтриаксон, подавлявший рост биопленок только 2 штаммов.
 529-534
529-534


Эпидемиология и молекулярно-генетическая характеристика возбудителей Лайм-боррелиоза, циркулирующих в популяции клещей на территории Алматинской области Республики Казахстан
Аннотация
Введение. Информация о географическом распространении различных видов комплекса Borrelia burgdorferi sensu lato (B. burgdorferi s.l.) имеет важное эпидемиологическое значение, поскольку различные геновиды связаны с определенными клиническими проявлениями Лайм-боррелиоза. Алматинская область Республики Казахстан считается эндемичной по клещевому боррелиозу, однако точная информация об уровне зараженности боррелиями клещей, обитающих в области, включая информацию о генотипах циркулирующих боррелий, отсутствует.
Целью данной работы было исследование клещей, снятых с людей в Алматинской области Республики Казахстан в 2018 г.
Материалы и методы. Клещей (n = 253) изучали на содержание в них ДНК B. burgdorferi s.l., генотипирование выявленных боррелий по участку гена 16S рРНК осуществляли методом секвенирования. Проведен анализ эпидемиологических данных по заболеваемости Лайм-боррелиозом в Алматинской области за 2013–2018 гг.
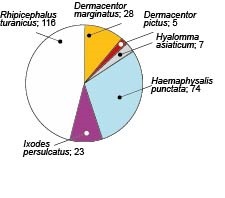
Результаты. Превалирующими видами клещей, снятых с людей, оказались Rhipicephalus turanicus (n = 116), Haemaphysalis punctata (n = 74), Dermacentor marginatus (n = 28) и Ixodes persulcatus (n = 23). Уровень зараженности клещей I. persulcatus бактериями B. burgdorferi s.l. составил 39,13% (n = 9). ДНК B. burgdorferi s.l. также была детектирована в единичных особях D. marginatus, H. punctata и R. turanicus, хотя данные виды не являются компетентными векторами B. burgdorferi s.l. В результате секвенирования положительных гомогенатов клещей были идентифицированы два генотипа B. burgdorferi s.l.: B. afzelii и B. garinii и/или B. bavariensis.
Выводы. На территории Алматинской области циркулируют по меньшей мере два геновида: B. afzelii и B. garinii и/или B. bavariensis.
 535-545
535-545


Анализ спорадических случаев инвазивного листериоза в мегаполисе
Аннотация
Введение. Листериоз — пищевая инфекция, наиболее опасная для лиц из групп риска. Восприимчивость к листерийной инфекции определяется комплексом причин: факторами окружающей среды, иммунитетом человека, вирулентностью микроорганизма. Усиливать восприимчивость к листериозу могут и ранее перенесенные инфекции, особенно вирусные, количество выявленных возбудителей которых регулярно возрастает.
Целью исследования была молекулярно-генетическая характеристика возбудителей спорадического инвазивного листериоза в мегаполисе, выделенных преимущественно в период роста заболеваемости гриппом и ОРВИ.
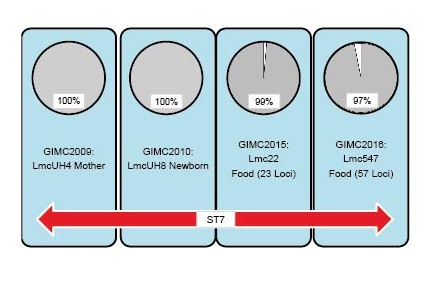
Материалы и методы. Изоляты Listeria monocytogenes были выделены от 18 госпитализированных пациентов в стационарах Москвы с ноября 2018 г. по октябрь 2019 г. В первой группе сравнения были изоляты из продуктов питания, а также изолят из рыбных пресервов. Во вторую группу сравнения вошли изоляты из окружающей среды, исследованные ранее. Клинические изоляты исследовали методами мультилокусного секвенирования, включающими стандартную схему MLST, дополненную локусами генов интерналинов. Полногеномное секвенирование с последующим анализом корового генома (cgMLST) применяли для сравнения изолятов аутохтонного генотипа (ST7).
Результаты. В случаях инвазивного листериоза 44% изолятов относилось к перинатальному листериозу, 27% составили изоляты от пациентов с менингитом. L. monocytogenes филогенетической линии II преобладала в этих группах заболеваний, случаи которых пришлись на период превышения эпидемического порога по гриппу в сезоне 2018/2019 гг. Листериозная пневмония, выявленная в самой старшей возрастной группе, была приурочена к сезону осенних ОРВИ и преимущественно вызвана L. monocytogenes филогенетической линии I. Исследование геномов изолятов ST7 показало идентичность коровых геномов бактерий, выделенных в паре родильница–новорожденный. Из пищевых изолятов ST7 наиболее близкородственным клиническим был изолят из мяса (23 локуса отличий, общая делеция в локусе MFS-транспортёра).Сопоставление перечня выявленных генотипов с данными европейских стран по анализу инвазивного листериоза показало, что для каждой страны характерен свой спектр генотипов, но ST7 был выявлен во всех рассмотренных выборках.
Выводы. Наряду с контролем производства и хранения продуктов питания своевременная вакцинация от сезонных респираторных инфекций, применение индивидуальных средств защиты в общественных местах могут снизить заболеваемость листериозом в группах риска.
 546-555
546-555


Изучение роли иммунитета к нейраминидазе вируса гриппа в защите от вторичной бактериальной пневмонии, индуцированной S. aureus после гриппозной инфекции у мышей
Аннотация
Введение. Пневмония является наиболее частым осложнением после гриппозной инфекции, с которым ассоциированы тяжелые случаи заболеваний и смертельные исходы во время сезонных и пандемических вспышек гриппа. Ранее мы показали, что вакцинирование мышей вирусоподобными частицами (ВПЧ), несущими гемагглютинин (НА) вируса гриппа, снижает смертность, вызванную бактериальными инфекциями после перенесенной гриппозной инфекции у мышей.
Цель данной работы — изучение возможности усиления защитного эффекта ВПЧ при дополнении их нейраминидазой (NA) вируса гриппа.
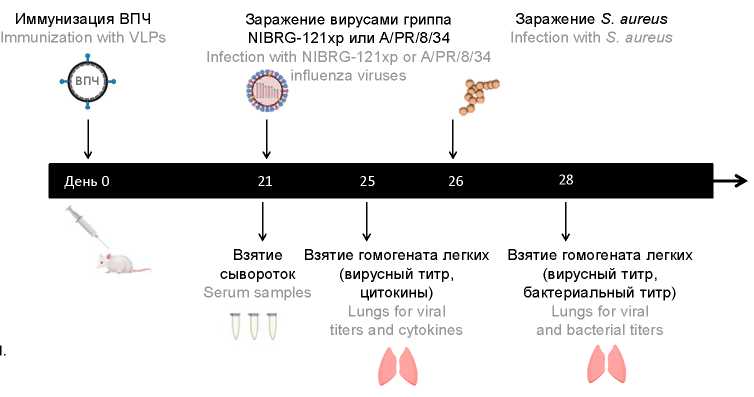
Материалы и методы. Изучали влияние Gag-ВПЧ, несущих HA или NA вируса гриппа A/Пуэрто-Рико/8/34, отдельно или в комбинации на модели вторичной бактериальной инфекции, индуцированной S. aureus после гриппозной инфекции гомологичным или гетерологичным вакцине вирусами гриппа.
Результаты. Коктейль HA-Gag-ВПЧ 100 нг + NA-Gag-ВПЧ 20 нг полностью предотвращал смертность, потерю веса и репликацию вируса, а также значительно снижал размножение бактерий в легких животных, зараженных гомологичным вирусом гриппа. Иммунизация этим же коктейлем защищала 60% животных от смертности, снижала потерю их веса и ингибировала размножение патогенов в легких животных с вторичной бактериальной инфекцией S. aureus после гриппозной инфекции гетерологичным вирусом гриппа H1N1, несмотря на отсутствие антител, ингибирующих НА и NA этого вируса.
Заключение. Наши результаты показывают, что парентеральная вакцинация ВПЧ, содержащими НА и NA, может улучшить исход вторичной бактериальной пневмонии, индуцированной S. aureus после гриппа, даже если вирус антигенно отличается от вакцинного штамма. При этом в нашей модели иммунитет к НА вируса гриппа имел превалирующее значение.
 564-577
564-577


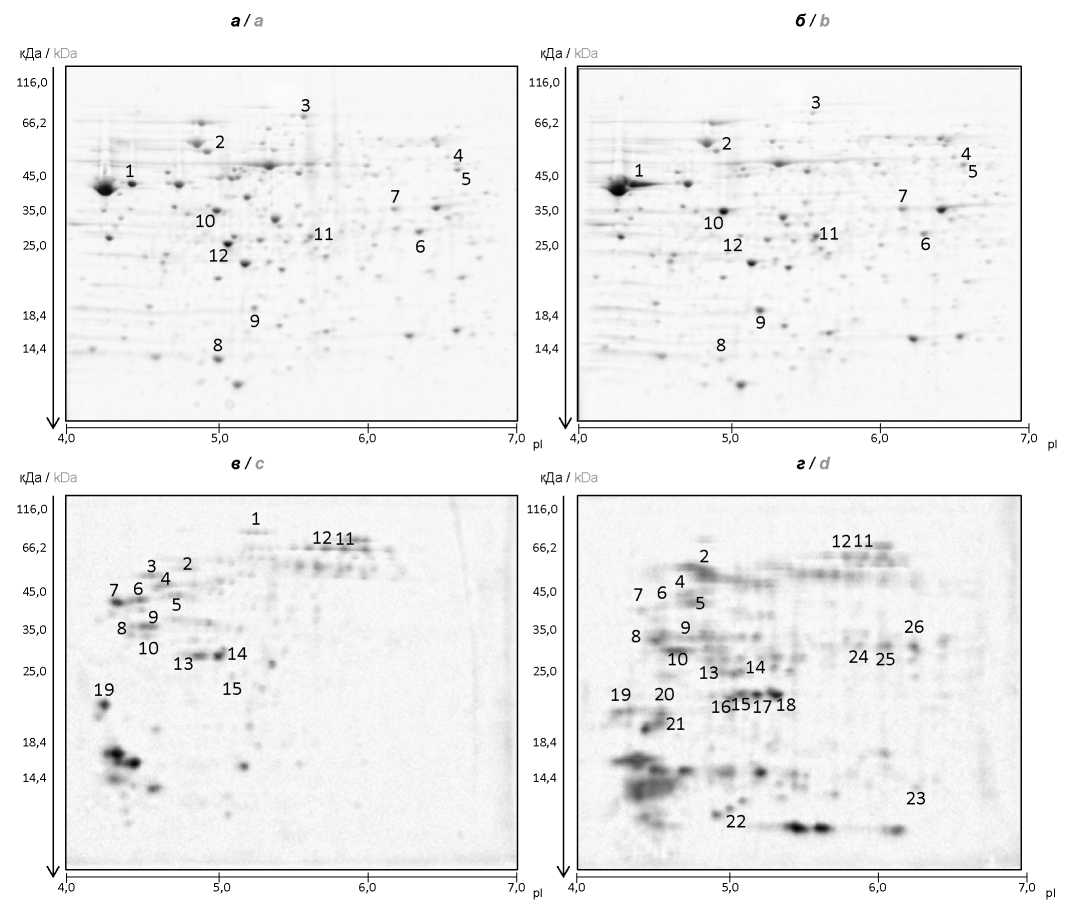
Протеомный анализ типичных и генетически измененных штаммов Vibrio cholerae О1 серогруппы биовара El Tor
Аннотация
Цель работы — сравнительное изучение экспрессии белков у типичных и генетически изменённых штаммов Vibrio cholerae O1 серогруппы биовара El Tor с помощью протеомного анализа.
Материалы и методы. В качестве модельных были использованы клинические штаммы V. cholerae — типичный M1062 (Астрахань, 1970) и генетически изменённый M1509 (Москва, 2012). Штаммы выращивали в LB бульоне рН 7,2. Получали фракции лизатов клеток и экзопротеинов и анализировали их методом 2D-электрофореза. Различающиеся белковые пятна исследовали масс-спектрометрически. Изучение выживаемости штаммов V. cholerae в условиях осмотического и оксидативного стресса проводили при инкубации штаммов в 3 М растворе NaCl или 20 мM растворе H2O2.
Результаты и обсуждение. При анализе лизатов клеток значительных отличий в экспрессии белков c известной функцией между изучаемыми штаммами не обнаружено. Подавляющая часть идентифицированных белков в лизатах функционально связана с углеводным обменом, метаболизмом аминокислот и энергетическими процессами в клетке. В то же время во фракции экзопротеинов геноварианта М1509 присутствовали в повышенных количествах белки (пероксидаза, супероксиддисмутаза, тиоредоксин, белки внешней мембраны OmpW и OmpT), защищающие клетки холерного вибриона от воздействия стрессовых факторов внешней среды. Последующее изучение устойчивости штаммов к осмотическому и оксидативному стрессу выявило лучшую выживаемость геноварианта при действии указанных факторов.
Заключение. Данные протеомного анализа типичного и генетически изменённого штаммов V. cholerae биовара El Tor свидетельствуют о повышенной экспрессии у геноварианта белков, обеспечивающих устойчивость вибрионов к действию стрессовых факторов внешней среды, что, возможно, является одной из причин их широкого распространения. Кроме того, полученные сведения позволят выявить новые биомаркеры, которые могут быть использованы для дифференциации типичных штаммов и геновариантов V. cholerae биовара El Tor.
 578-586
578-586


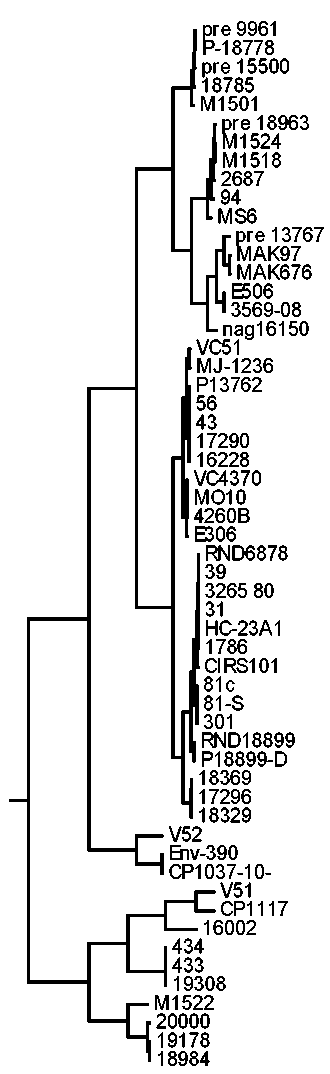
Совершенствование методики SNP-типирования штаммов Vibrio cholerae на основе анализа первичных данных полногеномного секвенирования
Аннотация
Цель работы — совершенствование метода оценки качества единичных нуклеотидных замен, используемых для SNP-типирования, на основе анализа их распределения в первичных данных полногеномного секвенирования (ридах).
Материалы и методы. В работе использованы данные полногеномного секвенирования 56 штаммов Vibrio cholerae, полученные на секвенаторах разных типов. Программное обеспечение разрабатывали на языке программирования Java. Кластерный анализ и построение дендрограммы проведены с использованием авторского программного обеспечения по методу UPGMA.
Результаты и обсуждение. Показана «нестабильность» определения ряда SNP в геноме возбудителя холеры. Разработан метод подбора перечня SNP для филогенетического анализа на основе обработки первичных данных полногеномного секвенирования (ридов). Предложена методика использования «контрольных геномов» при проведении кластерного анализа данных полногеномного секвенирования.
Заключение. Составлен перечень из 3198 «стабильных SNP» для проведения филогенетического анализа. Показана генетическая близость нетоксигенных штаммов, содержащих ген tcpA (ctxAB–tcpA+), и preCTX-штаммов V. cholerae.
 587-593
587-593


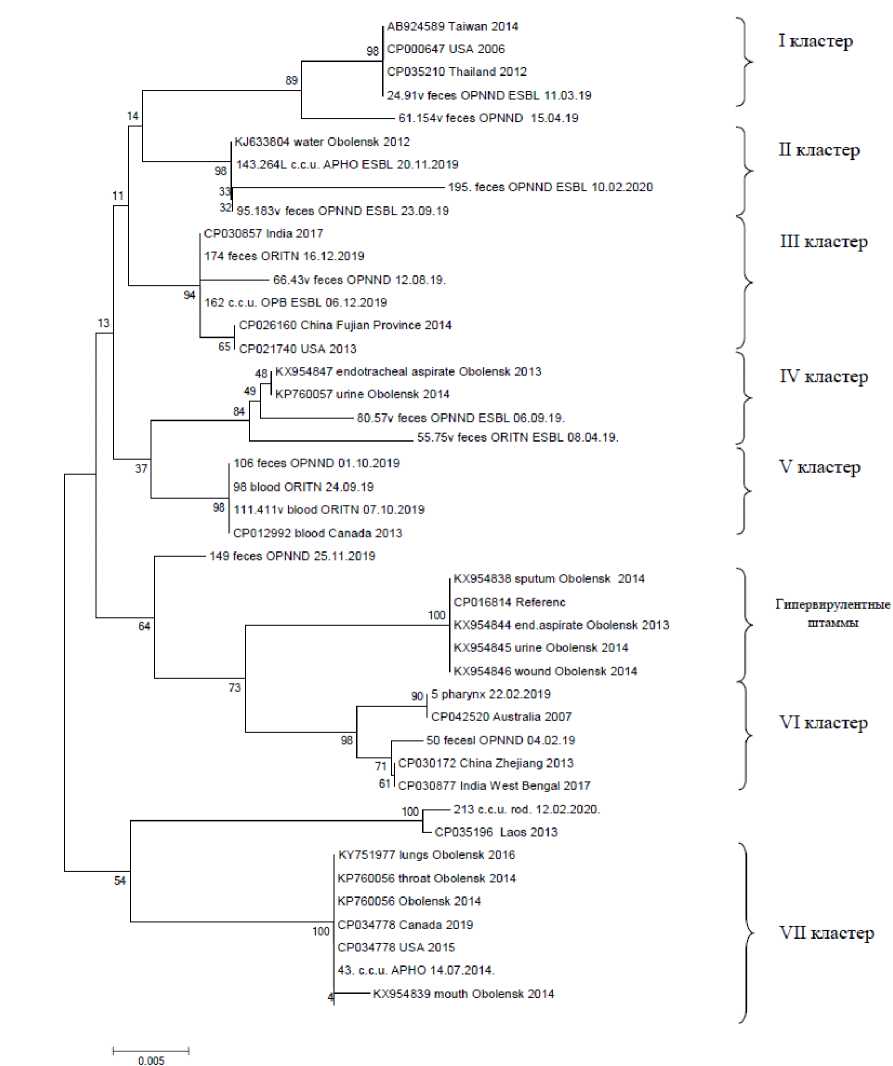
Филогенетический анализ родства штаммов Klebsiella pneumoniae по генам uge и fim
Аннотация
Введение. В настоящее время недостаточно данных о том, какова частота встречаемости штаммов Klebsiella pneumoniae с генами факторов вирулентности uge и fim среди женщин и новорожденных, что диктует необходимость проведения исследования распространенности K. pneumoniae (uge+, fim+) и определения степени гетерогенности выделенной популяции бактерий среди детей и взрослых.
Цель исследования — филогенетический анализ генов uge и fim штаммов K. pneumoniae для оценки гетерогенности изучаемой популяции и возможной кластеризации по локусу колонизации, временнóй и территориальной принадлежности.
Материалы и методы. Исследовано 65 штаммов K. pneumoniae, выделенных от 39 новорожденных и 24 женщин из проб фекалий, крови, мочи, последа, отделяемого цервикального канала, зева, шва. Две гемокультуры получены от одного пациента с интервалом в 2 нед, и 2 изолята выделены из отделяемого цервикального канала и шва одной пациентки. Детекцию наличия генов проводили методом полимеразной цепной реакции, нуклеотидные последовательности генов определяли секвенированием по Сэнгеру.
Результаты исследования. Частота встречаемости гена uge составила 53,8%, fim — 23,1%, что свидетельствует о большей распространенности в штаммах гена uge, чем fim (р < 0,001). Проведенный филогенетический анализ 18 нуклеотидных последовательностей гена uge и 4 нуклеотидных последовательностей гена fim показал, что штаммы распределились по 7 и 4 кластерам соответственно. Установлено, что как по гену uge, так и по fim нет четкой кластеризации по временнóй и территориальной принадлежности, возрасту пациента, биологическому материалу.
Обсуждение. Результаты филогенетического анализа демонстрируют генетическую гетерогенность изучаемой популяции K. pneumoniae, что подтверждено широкой географией и временем выявления наиболее генетически близких бактериальных изолятов.
 556-563
556-563


ОБЗОРЫ
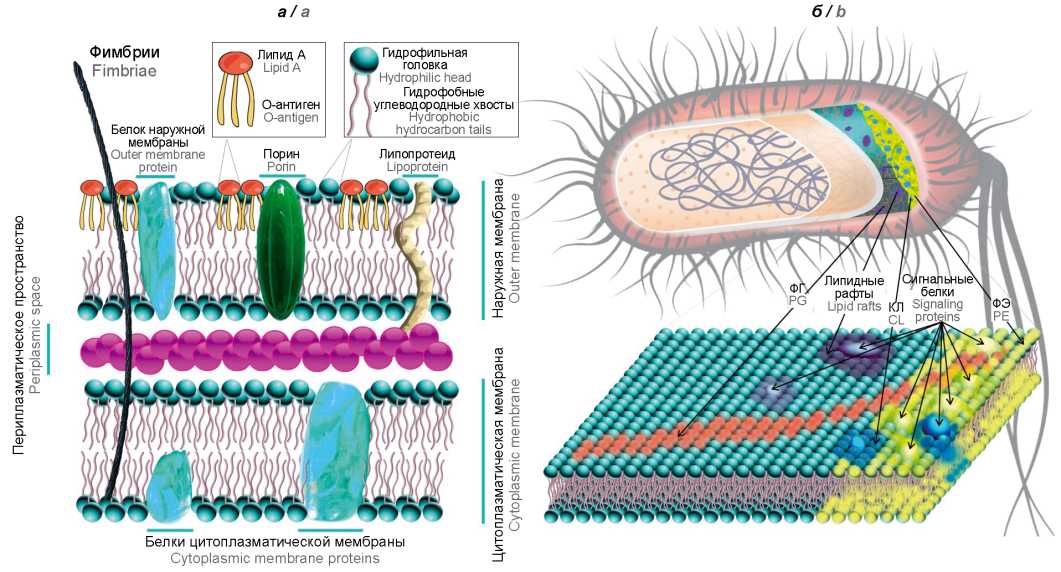
Значение мембранных фосфолипидов в реализации защитных стратегий бактерий
Аннотация
Для сохранения жизнеспособности в стрессовых условиях существования и реализации защитных стратегий бактерии должны воспринимать сигналы и быстро реагировать на экстремальные изменения параметров среды обитания. Результаты недавних экспериментальных исследований дополнили доминировавшую с 1970-х гг. парадигму о преимущественной роли фосфолипидов (ФЛ) как молекулярных строительных блоков для формирования клеточной стенки бактерий. Установлено, что специфические трансформации этих липидных доменов оказывают существенное влияние на форму и функционирование клеток, ремоделирование мембран и способность бактерий адаптироваться к стрессам окружающей среды. Физиологическая роль бактериальных фосфолипидов является плейотропной и определяет целостность и функцию клеток. Помимо ключевой структурной роли мембранных ФЛ в клетке, их промежуточные метаболиты способны выступать в качестве вторичных мессенджеров и играть важные сигнальные и регуляторные роли. Выявлено, что гомеостаз ФЛ имеет решающее значение для патогенеза бактериальных инфекций и необходим не только для поддержания жизнеспособности бактерий, но и для обеспечения их роста в период инфекции, а нарушение биосинтеза этих макромолекул снижает жизнеспособность бактерий. В последние десятилетия одним из главных достижений в концепции модели биологических мембран на основе «жидкой мозаики» стало понимание их доменной структуры. Это открытие представляет фундаментальный и практический интерес, поскольку фосфолипидные домены являются перспективной мишенью современных антимикробных стратегий. Цель настоящего обзора — обобщение современных представлений о структурной, метаболической и сигнальной роли мембранных ФЛ в реализации защитных механизмов бактерий и поддержании их жизнеспособности в неблагоприятных условиях среды обитания.
 594-603
594-603


КРАТКИЕ СООБЩЕНИЯ
Факторы, влияющие на смертность от новой коронавирусной инфекции в разных субъектах Российской Федерации
Аннотация
Актуальность. Влияние таких факторов, как плотность населения, практика тестирования на SARS-CoV-2 (в совокупности с самоизоляцией/карантином для инфицированных и контактных лиц) и температура воздуха, на распространение и смертность от COVID-19 в разных субъектах РФ недостаточно изучено.
Материалы и методы. Плотность населения в разных субъектах РФ оценивается как количество населения на 1 км 2 земли населенных пунктов; температура оценивается как среднее между температурами в январе и июле; практика тестирования на SARS-CoV-2 оценивается через коэффициент летальности (процент летальных случаев среди всех выявленных случаев COVID-19 с известным исходом (выздоровевших + умерших)) — при более активном тестировании выявляется больше случаев заболевания COVID-19 в легкой и средней форме и коэффициент летальности уменьшается, т.е. коэффициент летальности находится в обратной зависимости от активности тестирования.
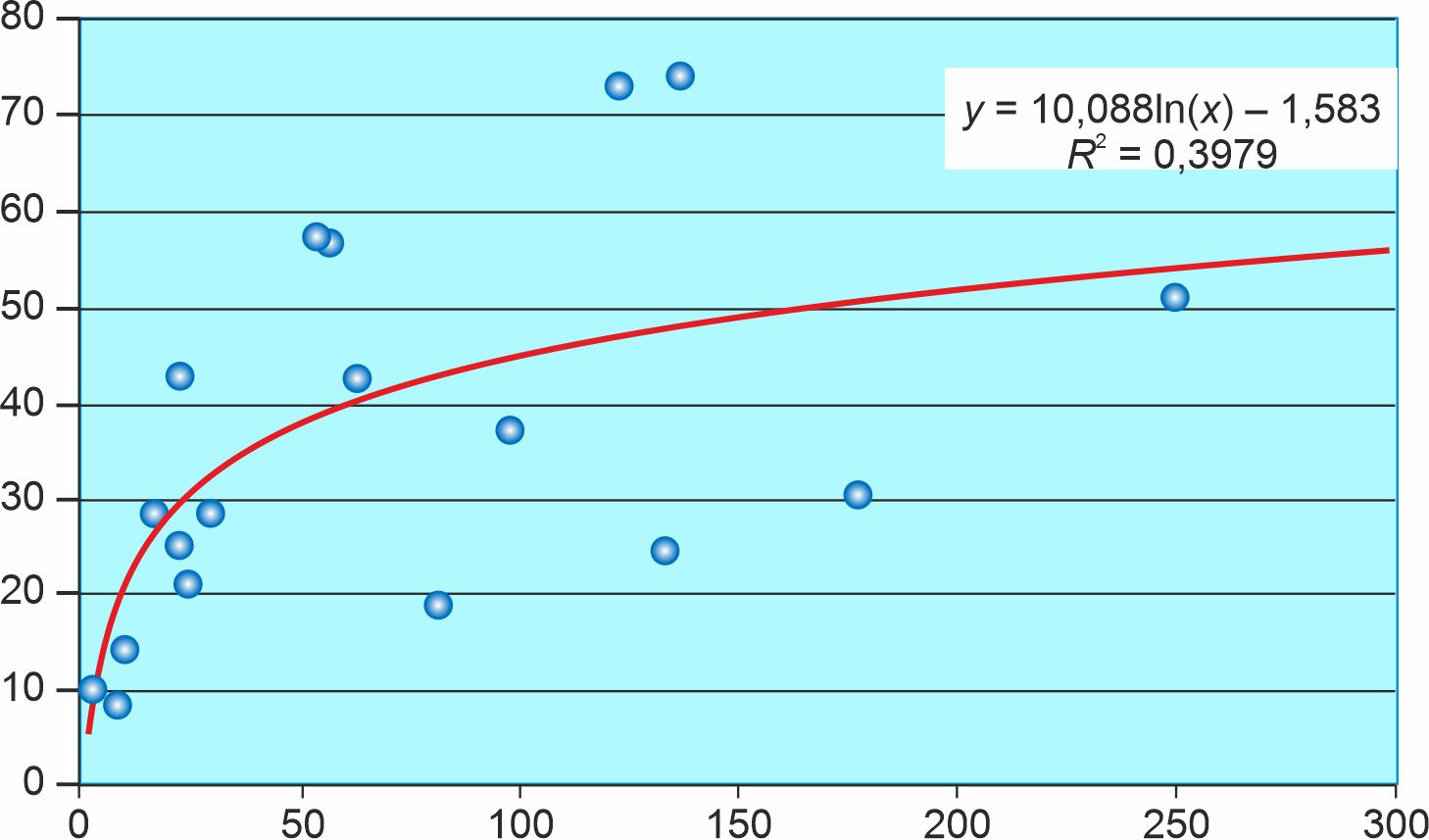
Результаты. Корреляция между плотностью населения и уровнем смертности от COVID-19 на 100 тыс. человек в 85 субъектах РФ на 22.11.2020 г. равна 0,53 (0,36; 0,67); корреляция между коэффициентом летальности и уровнем смертности — 0,62 (0,47; 0,74). Результаты линейной регрессии говорят о том, что плотность населения и коэффициент летальности положительно связаны с уровнем смертности от COVID-19 на 100 тыс. человек, а температура воздуха отрицательно связана с уровнем смертности от COVID-19 в 85 субъектах РФ.
Выводы. Более низкая плотность населения, более активное тестирование на SARS-CoV-2 и более высокая температура воздуха способствуют понижению уровня смертности от COVID-19 в разных субъектах РФ. В частности, следует принимать дополнительные меры для повышения уровня тестирования на SARS-CoV-2 среди разных категорий лиц, включая лиц, которые хотят тестироваться по собственной инициативе, лиц, обращающихся за медицинской помощью с симптомами ОРВИ, и контактных лиц для подтвержденных случаев COVID-19.
 604-607
604-607


ХРОНИКА
РЕЗОЛЮЦИЯ Всероссийской научно-практической интернет-конференции с международным участием «Молекулярная диагностика и биобезопасность — 2020» (Москва, 6–8 октября 2020 г.)
Аннотация
 608-609
608-609


РЕЗОЛЮЦИЯ Всероссийской научно-практической интернет-конференции с международным участием «Современная иммунопрофилактика: вызовы, возможности, перспективы» (Москва, 19–20 октября 2020 г.)
Аннотация
 610-612
610-612


РЕЗОЛЮЦИЯ 8-го Конгресса с международным участием «Контроль и профилактика инфекций, связанных с оказанием медицинской помощи (ИСМП-2020)» (Москва, 25–27 ноября 2020 г.)
Аннотация
 613-614
613-614


НЕКРОЛОГИ
 615-616
615-616


 617-620
617-620