Campylobacteriosis: genotypic characteristics of the pathogen and immunological status of patients
- Authors: Lobzin Y.V.1,2, Ermolenko K.D.1, Makarova M.A.2,3, Kaftyreva L.A.2,3, Martens E.A.1,2, Polev D.E.3, Ermolenko E.I.2,4
-
Affiliations:
- Pediatric Research and Clinical Center for Infectious Diseases
- I.I. Mechnikov North-Western State Medical University
- Saint-Petersburg Pasteur Institute
- Institute of Experimental Medicine
- Issue: Vol 101, No 3 (2024)
- Pages: 315-326
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18617
- DOI: https://doi.org/10.36233/0372-9311-531
- EDN: https://elibrary.ru/kihkmo
- ID: 18617
Cite item
Abstract
Introduction. Campylobacteriosis is among the leading causes of acute gastrointestinal infections. The severity of campylobacteriosis and the development of long-term complications may be influenced by the genotype of the pathogen, whose biological properties can affect immune response parameters.
The aim of the study was to identify common genotypes of epidemic clones of Campylobacter pathogens and to investigate characteristics of the immune response and severity of the disease.
Materials and methods. The study included 203 patients aged from 1 month to 17 years with campylobacteriosis who underwent treatment at the clinic of the Federal State Budgetary Institution "DNKCIB FMBA" in 2019–2021. The diagnosis was confirmed using polymerase chain reaction method. Patient samples were also analyzed using culture-based methods. Total DNA was extracted using the QIAamp DNA Mini Kit. Genetic determinants encoding virulence factors and MLST typing were performed using the ResFinder program. The immune status of patients was assessed on days 1 and 7 of the illness. Immunological investigation included measurement of serum immunoglobulin concentrations (IgA, IgM, IgG), C-reactive protein, and cytokines (IL-1β, IL-1, IL-2, IL-4, IL-5, IL-6, IL- 7, IL-8, IL-10, TNF-α, and IFN-γ).
Results. When analyzing the frequency of detection of Campylobacter sequence types in children with clinical intestinal infections, it was found that the profile of isolated isolates is most similar to those from countries of North America (USA and Canada), Northern Europe (Great Britain, Holland ) and Scandinavia (Denmark, Sweden, Finland). Identification of a pathogen with the flgE+, cdtA+, cdtC+ genotype was accompanied by a statistically significant increase in the level of IL-8 and a decrease in the content of IgA in the peripheral blood serum, which reflected the low efficiency of the immune response during infection with Campylobacter and predetermined the severe course of the infectious process during the disease.
Full Text
Introduction
Campylobacter spp. are one of the leading causative agents of acute intestinal infections (AII) in developed countries, exceeding in some regions the frequency of salmonellosis and escherichiosis registration. They are the cause of one third of cases of “travelers' diarrhea” among residents of industrial countries visiting regions with high levels of Campylobacter spp. circulation [1]. According to the latest WHO data, campylobacteriosis is one of the most common foodborne infectious diseases. Campylobacteriosis is being registered in all age groups, more often in children aged 1 to 5 years. Relative increase in disease cases is observed in older children and young people (compared to other age categories). Thermophilic Campylobacter species (TC) C. jejuni And C. coli [2] are of greatest importance in human infectious pathology, as they are characterized by a variety of genetic determinants that influence the pathogenetic and clinical features of the disease.
Unlike other pathogens of acute intestinal infections of a bacterial nature, thermophilic Campylobacter spp. are considered one of the most difficult microorganisms to cultivate, requiring special conditions and equipment. The isolation of a pure culture of the pathogen from stool samples for campylobacteriosis laboratory diagnosis is challenging due to their massive concomitant microbial contamination. In this regard, information on the incidence of this infection is fragmentary and does not provide a complete picture of its real spread [3, 4]. In recent years, the use of molecular research methods was considered not as an alternative, but as a mandatory addition to regulated diagnostic schemes for acute intestinal infections, allowing for the rapid and effective identification of pathogens of acute intestinal infections, including thermophilic Campylobacter spp. At the same time, it does not imply species identification and determination of sensitivity to antimicrobial drugs.
It is known that most Campylobacter spp. are resistant to the action of bile [5] and have the ability to colonize all parts of the intestine causing inflammation, edema, hyperplasia of the mucous membrane and erosions [6] . The pathogenic properties of campylobacteria are largely determined by their mobility, ability to adhere, invade and produce toxins. The flagella of campylobacteria determine their motility and movement along the epithelium [7, 8]. Adhesion and penetration of enterocytes contributes to the destruction of the intestinal mucosa, a pronounced inflammatory reaction and the development of hemorrhagic colitis [9]. Severe forms of campylobacteriosis are associated with the production of heat-stable and/or heat-labile enterotoxins and/or endotoxin (cell wall lipopolysaccharide), which affect the absorption of fluid and electrolytes, determining the development of diarrhea [10].
Campilobacter spp. genotyping methods are very important for epidemiological analysis allowing to identify “epidemic clones” — virulent strains capable of widespread distribution [11]. The study of the dominant genotypes of Campylobacter spp. can significantly supplement epidemiological monitoring, providing important information about the sources of infection, current transmission factors, and also assess the extent of the spread of resistance to antibacterial drugs [12]. It also cannot be ruled out that determining the genotype of the causative agent of campylobacteriosis may help clarify the prognosis of the severity of the infectious process and select the optimal drug therapy regimen [13].
Many researchers suggest that clinical manifestations of campylobacteriosis are largely due to the body's immune response [14]. Immunopathological reactions also predetermine numerous post-infectious complications, in particular, the development of Guillain–Barré syndrome, reactive arthritis and irritable bowel syndrome [15]. The variety of clinical forms and complications of campylobacteriosis determines special interest in the study of its pathogenesis, including the characteristics of the body’s immune response to this disease [16, 17]. At the same time, the severity of the infectious process and the formation of long-term complications can be determined by the genotype of the pathogen, the biological properties of which influence the parameters of the immune response [18]. That is why the features of the spread of epidemic clones, the association of the pathogen genotype with the severity of the disease and the immune response are of undoubted interest.
The purpose of this study was to detect the most common genotypes of epidemic campylobacteria clones and assess the nature of the immune response and the severity of the disease, taking into account the genotypic characteristics of pathogen.
Materials and methods
The study included 203 patients aged from 1 month to 17 years (mean age 4.8 ± 1.2 years) with a diagnosis of the underlying disease campylobacteriosis, treated in 2019–2021 at the PRCID clinic. The study was carried out with voluntary informed consent from the legal representatives of minor patients. The documentation and design of the study were approved at a meeting of the Local Ethics Committee at the FMBA (Protocol No. 11 of 03/05/2019).
To assess the severity of campylobacteriosis, the Clarke scale was used based on a score of the severity and duration of fever, diarrhea syndrome, vomiting and the general condition of the patient (Table 1). A score of more than 16 points corresponded to the severe form of campylobacteriosis.
Table 1. Clarke scale for assessing the severity of acute intestinal infections in children
Severity criterion | Severity of disease manifestations | ||
1 point | 2 points | 3 points | |
Number of bowel movements per day | 2–4 | 5–7 | > 8 |
Duration of diarrhea, days | 1–4 | 5–7 | > 8 |
Number of vomiting episodes per day | 1–3 | 4–6 | > 7 |
Duration of vomiting, days | 2 | 3–5 | > 6 |
Increase in body temperature, ºС | 37,1–38,2 | 38,3–38,7 | > 38,8 |
Duration of fever, days | 1–2 | 3–4 | > 5 |
Changes general state | Agitation or refusal to play | Lethargy or apathy | Convulsions or а loss of consciousness |
Duration of conservation pathological behavioral symptoms, days | 1–2 | 3–4 | > 5 |
The diagnosis of campylobacteriosis was confirmed based on the results of studies of stool samples using the polymerase chain reaction method with hybridization-fluorescence detection using the AmpliSens AII screen-FL reagent kit (Central Research Institute of Epidemiology of Rospotrebnadzor) for the detection and differentiation of DNA (RNA) of microorganisms of the genus Shigella spp./EIEC, Salmonella spp., Campylobacter spp. (thermophilic), Adenovirus (group F), Rotavirus (group A), Norovirus (genotype 2) and Astrovirus. Samples in which fluorescence levels were consistent with genetic determinants of thermophilic Campylobacter spp. were studied by the cultural method according to the Guidelines “Microbiological diagnosis of campylobacteriosis” No. 01/15702-8-34. To isolate Campylobacter strains spp., the following nutrient media were used: Columbia agar containing sheep blood (Sredoff LLC) and charcoal agar with a selective additive (Oxoid). Inoculation on nutrient media was carried out using cellulose acetate filters (Sartorius) with a pore diameter of 0.45 μm. Campylobacter cultivation was carried out under microaerophilic conditions at 42ºC for 48 hours using CO2GEN gas-generating packages (ThermoFisher).
For species identification, traditional routine tests were used based on the determination of key phenotypic characters: cell morphology and relation to Gram staining, cytochrome oxidase and catalase production, hydrolysis of sodium hippurate and indoxyl acetate, as well as MALDI-TOF mass spectrometry (Bruker Daltonik MALDI Biotyper).
Total DNA from bacterial strains was isolated using the QIAam p kit DNA Mini Kit (Qiagen). Whole-genome DNA sequencing was performed using the MiSeq (Illumina) and DNBSEQ-G50 (MGI) sequencing platforms with read lengths of 2×300 and 2×100. Raw readings were processed by Trim Galore v.0.6.7 program to remove adapter sequences and trim for quality. Processing quality control was carried out using the FastQC v.0.11.9 program. The genomes were assembled de novo using SPAdes assembler v.3.13.1 software. The assembly results were evaluated in QUAST v.5.2.0 program. The search for genetic determinants encoding virulence factors and MLST typing were carried out using the online resource platform Center for Genomic Epidemiology1.
The study of immune status was carried out on the 1st and 7th days of the disease. The immunological study included the quantitation of serum immunoglobulins (Ig) classes A, M, G, C-reactive protein, cytokines: interleukin (IL) -1β, -1, -2, -4, -5, -6, -7, -8, -10, tumor necrosis factor-α and interferon γ. The concentration of serum IgA, IgM, IgG and cytokines was assessed using an enzyme-linked immunosorbent assay (Vector-Best-Baltika). Fecal calprotectin (a non-invasive marker of neutrophilic intestinal inflammation) in stool samples was determined using a sandwich version of an enzyme-linked immunosorbent assay (R-Biopharm AG).
Statistical processing of the obtained data was carried out using the Statistica program for Windows v.10" (StatSoft). Quantitative indicators were assessed by compliance with the normal distribution using the Shapiro–Wilk criteria (for the number of subjects less than 50) and the Kolmogorov–Smirnov criterion (for the number of subjects more than 50). Quantitative indicators with a normal distribution were combined into variation series, in which arithmetic means (M) and standard deviations (SD) were calculated. Sets of quantitative indicators, the distribution of which differed from normal, were described using the values of the median and the lower and upper quartiles Me [Q1; Q3 ]. When comparing average values in normally distributed sets of quantitative data, the Student t-test was used; when comparing independent sets in cases where there were no signs of normal distribution of data, the Mann–Whitney U test was used.
Results
Clinical and laboratory data
The average severity of campylobacteriosis on the Clark scale was 12.6 ± 1.6 points. The severity of campylobacteriosis was assessed as moderate in 156 (76.85%) children, severe in 35 (17.24%), and mild in 12 (5.91%). Changes in the hemogram on the 1st day were characterized by neutrophilic leukocytosis in the range of 15–35 × 10 9 cells/l and acceleration of ESR in the range of 20–40 mm/h. During repeated studies on the 7th day, 16 (7.88%) patients still had minor deviations of hemogram parameters from normal values. An increase in the level of C-reactive protein was detected in 70.44% of patients. A strong positive correlation was established between the severity of campylobacteriosis, the total level of leukocytes (r = 0.56; p = 0.047) and C-reactive protein (r = 0.63; p = 0.016). Analysis of the levels of fecal calprotectin revealed a tendency towards its lower content in the blood serum of patients with a moderate form of the disease, compared to the severe form — 120.59 ± 47.21 and 242.80 ± 105.99 μg/g, respectively (p > 0.05).
Immunological status of patients with campylobacteriosis
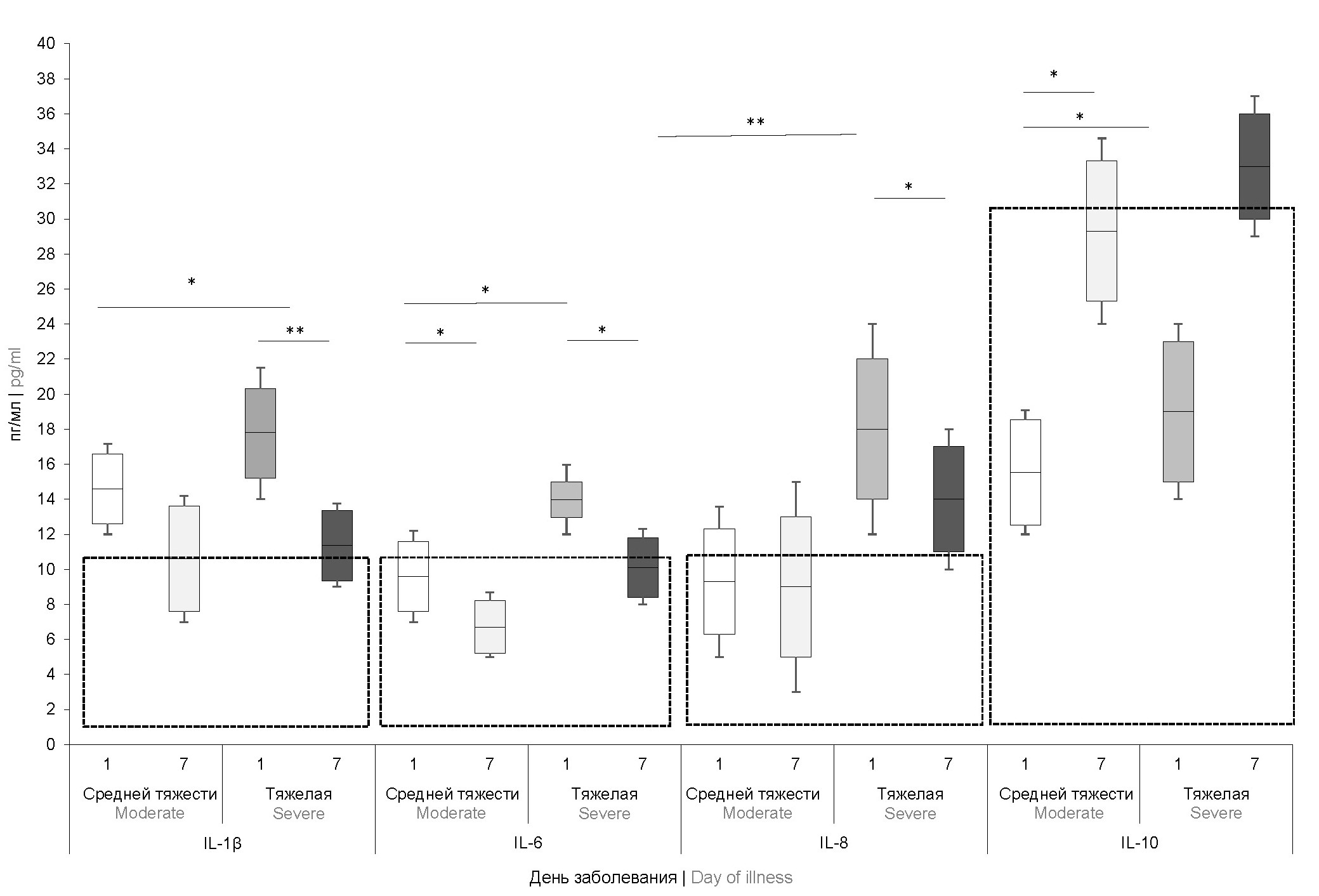
The study of cytokine status showed that the level of interferon-γ and tumor necrosis factor-α were within the reference values and did not differ significantly in patients with different degrees of campylobacteriosis severity. The most significant results were obtained when analyzing the pro-inflammatory cytokines IL-1β, -6, -8 and regulatory IL-10 (Fig. 1).
Fig. 1. Concentration of cytokines in the blood serum of children with moderate and severe forms of campylobacteriosis on days 1 and 7 of the disease (n = 42).
Here and in Fig. 2: * p < 0.05, ** p < 0.001. Reference values are highlighted by a frame.
On the 1st day of the disease, an increase in the level of IL-1β was detected in patients with moderate and severe forms of campylobacteriosis, IL-6 — only in patients with severe forms. The concentrations of these cytokines decreased by the 7th day of the disease, but only IL-6 levels reached normal reference values. Despite the fact that the level of IL-8 in a significant proportion of patients (44.1%) on the 1st day was within the reference values, there was a statistically significant increase in this indicator in patients with severe campylobacteriosis compared with the rest children (p = 0.002). An increase in this marker on the 1st day of the disease was one of the most significant predictors of severe campylobacteriosis (OR = 7.6 ± 1.7; p <0.001). A strong correlation was found between the level of IL-8 and the severity of campylobacteriosis (r = 0.781; p = 0.006). By the 7th day of the disease, this indicator decreased in all patients, but in severe cases of campylobacteriosis remained significantly higher than normal.
Fig. 2. Dynamics of the level of serum immunoglobulins in children with campylobacteriosis (n = 46).
The regulatory cytokine IL-10 in all patients on the 1st day of the disease did not exceed the reference values. At the same time, on the 7th day, this indicator significantly exceeded reference level in patients with severe campylobacteriosis, which can be explained by a compensatory reaction of the body aimed at preventing the development of allergic and autoimmune processes.
A study of the concentrations of Ig, IgM and IgG on the 1st day of the disease showed that, regardless of the severity of the infection, they were within the reference values. By day 7, there was an increase in the average level of immunoglobulins, statistically significant for IgA and IgM (p = 0.003 and p = 0.021, respectively). Both classes of immunoglobulins are produced during the acute period of the immune response, which appear in the blood upon first contact with infection.
Thermotolerant Campylobacter spp. were isolated by cultural method in 48 out of 203 studied stool samples from 28 children with gastroenteric and 20 with enterocolitic variants of campylobacteriosis, of which 6 (12.50%) had a mild disease, 30 (62.50%) had a moderate course and 12 ( 25.00%) — severe disease. Species identification revealed that 32 strains belonged to the species C. jejuni, 16 strains belonged to C. coli.
Frequency of detection of campylobacter virulence genes
Key virulence genes responsible for cheiotaxis, adhesion and colonization, invasion, morphogenesis of flagella and capsule, production of toxins and siderophores were present in all isolated strains of C. jejuni and C. coli (Table 2).
Table 2. Frequency of detection of virulence genes in Campylobacter strains spp.
Genes and factors virulence | C. jejuni (n = 32) | C. coli (n = 16) | p | Total (n = 48) | |||
n | % | n | % | n | % | ||
Mobility | |||||||
flaA | 32 | 100,00 | 16 | 100,00 | 0,05 | 48 | 100,00 |
flaB | 16 | 50,00 | 14 | 87,50 | 0,011 | 30 | 62,50 |
flhA | 28 | 87,50 | 12 | 75,00 | 0,05 | 40 | 83,33 |
flhB | 30 | 93,75 | 14 | 87,50 | 0,05 | 44 | 91,67 |
flgB | 32 | 100,00 | 16 | 100,00 | 0,05 | 48 | 100,00 |
flgE | 25 | 78,13 | 16 | 100,00 | 0,05 | 41 | 85,42 |
fliM | 32 | 100,00 | 16 | 100,00 | 0,05 | 48 | 100,00 |
fliY | 32 | 100,00 | 16 | 100,00 | 0,05 | 48 | 100,00 |
Adhesion | |||||||
cadF | 29 | 90,63 | 12 | 75,00 | 0,05 | 41 | 85,42 |
dnaJ | 32 | 100,00 | 16 | 100,00 | 0,05 | 48 | 100,00 |
jlpA | 20 | 62,50 | 7 | 43,75 | 0,05 | 27 | 56,25 |
pldA | 22 | 68,75 | 7 | 43,75 | 0,05 | 29 | 60,42 |
racR | 28 | 87,50 | 12 | 75,00 | 0,05 | 40 | 83,33 |
virB11 | 11 | 34,38 | 5 | 31,25 | 0,05 | 16 | 33,33 |
Chemotaxis | |||||||
cheA | 30 | 93,75 | 16 | 100,00 | 0,05 | 46 | 95,83 |
cheB | 31 | 96,88 | 13 | 81,25 | 0,05 | 44 | 91,67 |
cheR | 24 | 75,00 | 12 | 75,00 | 0,05 | 36 | 75,00 |
cheW | 30 | 93,75 | 10 | 62,50 | 0,0062 | 40 | 83,33 |
cheY | 31 | 96,88 | 15 | 93,75 | 0,05 | 46 | 95,83 |
cheZ | 28 | 87,50 | 9 | 56,25 | 0,015 | 37 | 77,08 |
Infestation | |||||||
iamA | 29 | 90,63 | 15 | 93,75 | 0,05 | 44 | 91,67 |
ciaB | 29 | 90,63 | 13 | 81,25 | 0,05 | 42 | 87,50 |
ceuE | 24 | 75,00 | 16 | 100,00 | 0,05 | 40 | 83,33 |
Toxins | |||||||
cdtA | 20 | 62,50 | 10 | 55,56 | 0,05 | 30 | 62,50 |
cdtB | 32 | 100,00 | 16 | 100,00 | 0,05 | 48 | 100,00 |
cdtC | 21 | 65,63 | 12 | 75,00 | 0,05 | 33 | 68,75 |
wlaN | 4 | 12,50 | 2 | 12,50 | 0,05 | 6 | 12,50 |
Capsule | |||||||
kpsM | 29 | 90,63 | 13 | 81,25 | 0,05 | 42 | 87,50 |
Siderophores | |||||||
cfrA | 27 | 84,38 | 11 | 68,75 | 0,05 | 38 | 79,17 |
Fur | 30 | 93,75 | 14 | 87,50 | 0,05 | 44 | 91,67 |
Analysis of genetic determinants of pathogenicity showed that genes associated with motility (flaA, flaB, flhA, flhB, flgB, flgE, fliM, fliY), adhesion (cadF, dnaJ, jlpA, pldA, racR, virB11), colonization (cheA, cheB, cheR, cheW, cheY, cheZ), invasion (iamA, ciaB, ceu E), synthesis of toxins (cdtA, cdtB, cdtC, wlaN), capsule (kpsM) and siderophores (cfrA, Fur) without statistically significant differences were found in C. jejuni and C. coli strains.
The prevalence of genes encoding flagellin synthesis, according to the total data, ranged from 62.50% (flaB) to 100% (flaA, flgB, fliM, fliY). Analysis of the results of detection of genes associated with adhesion showed that all strains contained the dna J gene. No statistical differences in the presence of genetic determinants encoding the ability to chemotaxis were detected. All strains were characterized by the presence of the cdtB gene, responsible for the production of a cytolethal toxin involved in the suppression of the proliferation of enterocytes with their subsequent death. Invasion-associated genes iamA, ciaB, ceuE were identified in 91.67, 87.50, 83.33% of Campylobacter strains spp., respectively. Analysis of the presence of genetic virulence determinants encoding the synthesis of the capsule and siderophores did not reveal significant differences in the strains of C. jejuni and C. coli.
Campylobacter spp. characterized by virulence genotype flgE+, cdtA+ and cdtC+ was found in 83.3% of patients with severe campylobacteriosis. According to multivariate analysis, this genotype increases the likelihood of severe campylobacteriosis by 12.57 [3.159; 50.019] times (p < 0.001).
Sequence types Campylobacters and their geographical distribution
Phylogenetic analysis showed that some strains classified as C. jejuni formed a separate genetically distinct group. In 8 strains, some alleles belonging to the C. jejuni taxonomic cluster were also found in C. coli and vice versa, caused by the genetic mosaic occurring within the genus.
A comparison of the diversity of constitutional genes in C. coli and C. jejuni was performed. Among 32 C. jejuni strains, multilocus sequencing typing (MLST) revealed 18 different MLST sequence types, which were sorted into 12 different complexes. The geographical distribution of the identified genotypes in other regions of the world is presented in Table. 3.
Table 3. Distribution of sequence types and core -genomic sequence types C. jejuni in various countries (n = 32)
Strain number | Sequence type | Denmark | UK | Holland | Australia | USA | Canada | Spain | Norway | Luxembourg | Japan | Uruguay | China | Belgium | France | Finland | Sweden | Czech Republic |
EI0796 | 21 | + | + | + | + | + | + | + | + | + | + | + | + | + | – | – | – | – |
EI0797 | 137 | – | + | + | – | + | + | – | – | + | – | – | – | – | + | – | – | + |
EI0798 | 38 | – | + | + | – | + | + | – | – | + | – | + | – | – | – | – | – | – |
EI0800 | 48 | + | + | + | + | + | + | + | – | + | – | + | – | – | + | + | + | + |
EI0801 | 3503 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
EI0802 | 49 | + | + | + | – | – | – | – | – | + | – | – | + | – | – | + | – | – |
EI0803 | 52 | – | + | + | + | – | + | – | – | – | – | + | + | + | – | – | – | – |
EI0804 | 2100 | – | + | + | – | – | – | – | – | – | – | – | – | – | – | + | – | – |
EI0805 | 61 | – | + | + | + | + | + | – | – | + | – | + | – | + | – | – | – | – |
EI0806 | 122 | – | + | + | – | + | – | – | – | + | – | – | – | + | – | – | – | – |
EI0807 | 206 | – | + | + | – | – | – | – | – | + | + | – | – | – | – | + | – | – |
EI0808 | 353 | – | + | + | – | + | + | – | – | + | – | – | – | + | – | – | – | – |
EI0809 | 524 | – | – | + | + | – | – | – | – | – | – | – | – | – | – | – | + | – |
EI0810 | 354 | – | + | + | + | + | + | – | – | + | + | + | – | – | + | – | – | – |
EI0811 | 443 | – | + | + | – | – | + | – | – | + | – | – | + | – | + | – | – | – |
EI0812 | 584 | – | + | + | – | – | – | – | – | + | – | – | – | – | – | – | – | – |
EI0813 | 824 | – | + | + | – | – | – | + | – | + | + | – | – | + | – | – | – | – |
EI0814 | 305 | – | + | + | – | – | – | + | – | – | – | + | – | – | – | – | – | – |
Note. «+» — presence; «‒» — absence of C. jejuni in a given country.
When matching sequence types of Campylobacters isolated from patients with data from international databases, the similar sequence types were shown to be most common in North America ( USA and Canada), Northern Europe (Great Britain, Holland) and Scandinavia (Denmark, Sweden, Finland). Among 16 C. coli strains, 10 different types of MLST sequences were detected. Various types of sequences are maximally represented in Great Britain, Holland and Luxembourg. The nature of the geographical distribution of isolated C. jejuni and C. coli in patients in the study had a number of similarities, in particular, the dominance of sequence types, most common in Northern Europe, was noted. Close socio-economic ties and geographic proximity of the countries represented create the prerequisites for the multiple routes of the pathogen transmission, which probably explains such a distribution.
Discussion
Contradictions and lack of consistency in data on the forms of the disease and severity are associated with the pathogenic potential of the pathogen and differences in the immune response. Previous studies have demonstrated the high conservation of flagellar genes, which are critical virulence factors [19, 20] ensuring the colonization and survival of campylobacteria [21]. Flagella are necessary to resist intestinal motility, which might otherwise displace the microorganism from the gastrointestinal tract [22]. The flagellar filament consists of the protein flagellin, which is encoded by two adjacent genes: flaA and flaB. In the works of L. Koolman et al. flaB-negative campylobacters have been shown to exhibit partial motility and can remain viable [23]. However, in a number of other studies there was evidence that the absence of flaB reduced the colonization ability and virulence of the pathogen [24]. It is also worth noting that the absence of flaB was significantly more often detected in C. jejuni isolates compared to C. coli.
Genes flaA and flaB involved in the assembly of flagella, according to L. Koolman et al., are critical for invasion [25]. Not a single strain that lacked both genes simultaneously was isolated in this study, which partially confirms this hypothesis.
When analyzing the frequency of detection of virulence genes, attention was drawn to the rare detection of the virB11 and wlaN genes. The virB11 gene encodes proteins responsible for adhesion to enterocytes [25]. According to D. Bacon et al., identification of the vir B11 gene led to a sharp increase in the ability of adhesion compared to wild-type strains [26]. Similarly, the wlaN gene that encodes a b-1,3-galactosyltransferase involved in cell wall synthesis, greatly increases the ability of campylobacters to attach to the surface of the intestinal epithelium [27]. Apparently, the frequent detection of isolates that do not contain these genes indicates their auxiliary role, and their absence does not lead to a significant decrease in virulence.
Gene wla is considered in a number of publications as a key trigger of immunopathological reactions, in particular, triggering the development of autoimmune polyradiculopathies [28]. Its low prevalence in the study group may partly explain the lack of a large number of reports of confirmed cases of Guillain–Barré syndrome in children after campylobacteriosis in Russia.
The catalase gene kat A was detected in 78% of isolates. This gene helps protect Campylobacter spp. against oxidative stress, increases survival within macrophages. At the same time, the presence of this gene leads to increased resistance to antibacterial drugs [29]. Iron uptake genes (cfrA and fur) were also present in the majority of isolates (79.17 and 91.67%, respectively).
In the group with severe campylobacteriosis, the IgA level was statistically significantly lower than in moderate campylobacteriosis (p = 0.239). It cannot be excluded that IgA deficiency negatively affects the course of the disease due to insufficient neutralization of pathogen toxins. An increase in the level of IgG, which makes up 75–80% of antibodies in plasma, providing long-term humoral protection against re-infection, was observed only in patients with severe campylobacteriosis (p = 0.039). Obviously, in these patients, the elimination of Campylobacter from the body was delayed, which led to a longer and more massive stimulation of the immune response by pathogen antigens. Another predictor of severe campylobacteriosis was an increase in the level of IL-8. IL-8 is considered as a key trigger of nonspecific immune defense, being a chemoattractant affecting mainly neutrophils and monocytes [14]. An assessment of the dynamics of IL-8 levels also demonstrated undoubted prognostic significance. Long-term persistence of an increase in this marker acted as an unfavorable prognostic factor and was often observed during a protracted course of the disease. It was noteworthy that the pathogen genotype flgE+, cdtA+, cdtC+ was more often detected in severe campylobacteriosis in the presence of significant deviations in the parameters of the immune response, which may be an additional prerequisite for the severe course of the disease. Analysis of the immunological parameters of the course of campylobacteriosis with the causative agent genotype flgE+, cdtA+, cdtC+ revealed statistically significant differences in the level of IL-8 (0.013) and IgA (p = 0.021) on the 1st day of the disease compared to patients, in which other genotypes of pathogens were detected.
Conclusion
Thus, when analyzing the frequency of detection of campylobacter sequence-types in children with clinical intestinal infections, it was found that the profile of isolated isolates is most similar to those from countries of North America (USA and Canada), Northern Europe (Great Britain, Holland) and Scandinavia (Denmark, Sweden, Finland). Children with C. coli and C. jejuni infections with genotype flgE+, cdtA+, cdtC+, had a number of clinical differences during the infectious process from patients with other genotypes of the pathogen. Identification of Campilobacter spp. with the genotype flgE+, cdtA+, cdtC+ on the 1st day of the disease is accompanied by a more frequent increase in the level of IL-8 and a decrease in the content of IgA in the peripheral blood serum, which may indicate insufficient efficiency of the immune response when infected with campylobacters of this genotype. It cannot be excluded that the identified features of the immune response during infection with campylobacters with genotypes flgE+, cdtA+, cdtC+ underlie the long-term persistence of disease symptoms and an increase in the severity of the disease. Considering the high social and clinical significance of campylobacteriosis, it is advisable to further study the genotypic composition of circulating pathogens to assess the risk of developing severe forms of the disease and the formation of its long-term complications.
1 Center for Genomic Epidemiology. URL: http://www.genomicepidemiology.org/services
About the authors
Yury V. Lobzin
Pediatric Research and Clinical Center for Infectious Diseases; I.I. Mechnikov North-Western State Medical University
Email: ermolenko.kd@yandex.ru
ORCID iD: 0000-0002-6934-2223
D. Sci. (Med.), Professor, RAS Full Member, President, Pediatric Research and Clinical Center for Infectious Diseases; Head, Department of infection diseases, I.I. Mechnikov North-Western State Medical University
Russian Federation, St. Petersburg; St. PetersburgKonstantin D. Ermolenko
Pediatric Research and Clinical Center for Infectious Diseases
Email: ermolenko.kd@yandex.ru
ORCID iD: 0000-0002-1730-8576
Cand. Sci. (Med.), Head, Research department of intestinal infections
Russian Federation, St. PetersburgMaria A. Makarova
I.I. Mechnikov North-Western State Medical University; Saint-Petersburg Pasteur Institute
Email: ermolenko.kd@yandex.ru
ORCID iD: 0000-0003-3600-2377
D. Sci. (Med.), leading researcher, Head, Department of intestinal infections, Pediatric Research and Clinical Center for Infectious Diseases; Associate Professor, Department of medical microbiology, I.I. Mechnikov North-Western State Medical University
Russian Federation, St. Petersburg; St. PetersburgLidia A. Kaftyreva
I.I. Mechnikov North-Western State Medical University; Saint-Petersburg Pasteur Institute
Email: ermolenko.kd@yandex.ru
ORCID iD: 0000-0003-0989-1404
D. Sci. (Med.), leading researcher, Typhoid epidemiology group, Saint-Petersburg Pasteur Institute; Professor, Department of medical microbiology, I.I. Mechnikov North-Western State Medical University
Russian Federation, St. Petersburg; St. PetersburgElvira A. Martens
Pediatric Research and Clinical Center for Infectious Diseases; I.I. Mechnikov North-Western State Medical University
Email: ermolenko.kd@yandex.ru
ORCID iD: 0000-0001-6093-7493
Cand. Sci. (Med.), Head, Clinical diagnostic laboratory, Pediatric Research and Clinical Center for Infectious Diseases; Assistant Professor, Department of medical microbiology, North-Western State Medical University named after I.I. Mechnikov
Russian Federation, St. Petersburg; St. PetersburgDmitry E. Polev
Saint-Petersburg Pasteur Institute
Author for correspondence.
Email: ermolenko.kd@yandex.ru
ORCID iD: 0000-0001-9679-2791
Cand. Sci. (Biol.), senior researcher, Metagenomic research group
Russian Federation, St. PetersburgElena I. Ermolenko
I.I. Mechnikov North-Western State Medical University; Institute of Experimental Medicine
Email: ermolenko.kd@yandex.ru
ORCID iD: 0000-0002-2569-6660
D. Sci. (Med.)., Head, Laboratory of molecular microbiology, Institute of Experimental Medicine; Professor, Department of medical microbiology, I.I. Mechnikov North-Western State Medical University
Russian Federation, St. Petersburg; St. PetersburgReferences
- Hameed A., Woodacre A., Machado L.R., Marsden G.L. An updated classification system and review of the lipooligosaccharide biosynthesis gene locus in Campylobacter jejuni. Front. Microbiol. 2020;11:677. DOI: https://doi.org/10.3389/fmicb.2020.00677
- Потапова Т., Ермоленко К., Холин А. и др. Заболеваемость острыми кишечными инфекциями в Санкт-Петербурге на фоне пандемии COVID-19. Журнал инфектологии. 2022; 14(3):37–44. Potapova T.V., Ermolenko K.D., Kholin A., et al. Incidence of acute intestinal infections in Saint Petersburg during COVID-19 pandemic. DOI: https://doi.org/10.22625/2072-6732-2022-14-3-37-44 EDN: https://elibrary.ru/kikypp
- Kaakoush N.O., Castaño-Rodríguez N., Mitchell H.M., Man S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015;28(3):687–720. DOI: https://doi.org/10.1128/cmr.00006-15
- Климова О., Гончар Н., Раздьяконова И., Лобзин Ю. Этиологические и эпидемиологические особенности инфекционных гемоколитов у госпитализированных пациентов детского возраста. Журнал инфектологии. 2021;13(1):86–92. Klimova O.I., Gonchar N.V., Razd'yakonova I.V., Lobzin Yu.V. Etiological and epidemiological characteristics of infectious hemocolitis in hospitalized pediatric patients. Journal Infectology. 2021;13(1):86–92. DOI: https://doi.org/10.22625/2072-6732-2021-13-1-86-92 EDN: https://elibrary.ru/jxnncq
- Жданов К.В., Захаренко С.М., Львов Н.И., Козлов К.В. Противодействие инфекциям в эпоху современных угроз. Инфекционные болезни: Новости. Мнения. Обучение. 2017; (6):85–91. Zhdanov K.V., Zakharenko S.M., Lvov N.I., Kozlov K.V. Counteracting infections in the age of current threats. Infectious Diseases: News, Views, Education. 2017;(6): 85–91. EDN: https://elibrary.ru/zvghkz
- Sher A.A., Ashraf M.A., Mustafa B.E., Raza M.M. Epidemiological trends of foodborne Campylobacter outbreaks in the United States of America, 1998–2016. Food Microbiology. 2021;97:103751. DOI: https://doi.org/10.1016/j.fm.2021.103751
- Dos Santos F.M.S., Low K.H., Chai L.C. Thermophilic and non-thermophilic Campylobacter species emits distinct volatile organic compounds in different culture media and growth phases. Res. Square. 2022. Preprint. DOI: https://doi.org/10.21203/rs.3.rs-1247479/v1
- Callahan S.M., Dolislager C.G., Johnson J.G. The host cellular immune response to infection by Campylobacter spp. and its role in disease. Infect. Immun. 2021;89(8):e0011621. DOI: https://doi.org/10.1128/iai.00116-21
- Kemper L., Hensel A. Campylobacter jejuni: targeting host cells, adhesion, invasion, and survival. Appl. Microbiol. Biotechnol. 2023;107(9):2725–54. DOI: https://doi.org/10.1007/s00253-023-12456-w
- Tegtmeyer N., Sharafutdinov I., Harrer A., et al. Campylobacter virulence factors and molecular host–pathogen interactions. Curr. Top. Microbiol. Immunol. 2021;431:169–202. DOI: https://doi.org/10.1007/978-3-030-65481-8_7
- Wassenaar T.M. Toxin production by Campylobacter spp. Clin. Microbiol. Rev. 1997;10(3):466–76. DOI: https://doi.org/10.1128/cmr.10.3.466
- Lopes G.V., Ramires T., Kleinubing N.R., et al. Virulence factors of foodborne pathogen Campylobacter jejuni. Microb. Pathog. 2021;161(Pt. A):105265.13. DOI: https://doi.org/10.1016/j.micpath.2021.105265
- Newby T.J. Protective immune responses in the intestinal tract. In: Newby T.J., Stokes C.R. Local Immune Responses of the Gut. Boca Raton;2019:143–98. DOI: https://doi.org/10.1201/9780429279508
- Жданов К., Яременко М., Финогеев Ю., Захаренко С. Иммуно-патогенетические аспекты лихорадки у инфекционных больных. Журнал инфектологии. 2014;5(1):5–17. Zhdanov K.V., Yaremenko M.V., Finogeev Yu.P., Zakharenko S.M. Clinical and pathogenetic aspects of fever in patients with infectious diseases. Journal Infectology. 2014;5(1):5–17. EDN: https://elibrary.ru/redmqr
- Goni M., Muhammad I., Goje M., et al. Campylobacter in dogs and cats; its detection and public health significance: а review. Adv. Anim. Vet. Sci. 2017;5(6):239–48. DOI: https://doi.org/10.17582/journal.aavs/2017/5.6.239.248
- Савиных М.В., Калужских Т.И., Савиных Н.А., Егорова Т.В. Клинико-эпидемиологические аспекты сальмонеллеза и кампилобактериоза у детей. Журнал инфектологии. 2020;12(4 S1):97. Savinykh M.V., Kaluzhskikh T.I., Savinykh N.A., Egorova T.V. Clinical and epidemiological aspects of salmonellosis and campylobacteriosis in children. Journal Infectology. 2020;12(4 S1):97. EDN: https://elibrary.ru/ruwekh
- Горелов А.В. Кампилобактериоз у детей. Инфекционные болезни. 2004;2(3):80–2. Gorelov A.V. Campilobacteriosis in children. Infectious Diseases. 2004;2(3):80–2. EDN: https://elibrary.ru/iadjkj
- Shahreza M.S., Dehkordi N.G., Nassar M.F., Al-Saedi R. Genotyping of Campylobacter jejuni isolates from raw meat of animal species. Acad. J. Health Sci. Medicina Balear. 2022;47(4):52–7.
- Strakova N., Michova H., Shagieva E., et al. Genotyping of Campylobacter jejuni and prediction tools of its antimicrobial resistance. Folia Microbiol. (Praha). 2024;69(1):207–19. DOI: https://doi.org/10.1007/s12223-023-01093-5
- Peters S., Pascoe B., Wu Z., et al. Campylobacter jejuni genotypes are associated with post-infection irritable bowel syndrome in humans. Commun. Biol. 2021;4(1):1015. DOI: https://doi.org/10.1038/s42003-021-02554-8
- Datta S., Niwa H., Itoh K. Prevalence of 11 pathogenic genes of Campylobacter jejuni by PCR in strains isolated from humans, poultry meat and broiler and bovine faeces. J. Med. Microbiol. 2003;52(Pt. 4):345–8. DOI: https://doi.org/10.1099/jmm.0.05056-0
- Müller J., Schulze F., Müller W., Hänel I. PCR detection of virulence-associated genes in Campylobacter jejuni strains with differential ability to invade Caco-2 cells and to colonize the chick gut. Vet. Microbiol. 2006;113(1-2):123–9. DOI: https://doi.org/10.1016/j.vetmic.2005.10.029
- Jones M.A., Marston K.L., Woodall C.A., et al. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 2004;72(7):3769–76. DOI: https://doi.org/10.1128/iai.72.7.3769-3776.2004
- Hendrixson D.R., DiRita V.J. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 2004;52(2):471–84. DOI: https://doi.org/10.1111/j.1365-2958.2004.03988.x
- Koolman L., Whyte P., Burgess C., Bolton D. Virulence gene expression, adhesion and invasion of Campylobacter jejuni exposed to oxidative stress (H2O2). Int. J. Food Microbiol. 2016;220:33–8. DOI: https://doi.org/10.1016/j.ijfoodmicro.2016.01.002
- Konkel M.E., Klena J.D., Rivera-Amill V., et al. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 2004;186(11):3296–303. DOI: https://doi.org/10.1128/jb.186.11.3296-3303.2004
- Koolman L., Whyte P., Burgess C., Bolton D. Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 2015;12(5):424–32. DOI: https://doi.org/10.1089/fpd.2014.1883
- Bacon D.J., Alm R.A., Burr D.H., et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 2000;68(8):4384–90. DOI: https://doi.org/10.1128/iai.68.8.4384-4390.2000
- Talukder K.A., Aslam M., Islam Z., et al. Prevalence of virulence genes and cytolethal distending toxin production in Campylobacter jejuni isolates from diarrheal patients in Bangladesh. J. Clin. Microbiol. 2008;46(4):1485–8. DOI: https://doi.org/10.1128/jcm.01912-07
- Guirado P., Paytubi S., Miró E., et al. Differential distribution of the wlaN and cgtB genes, associated with Guillain–Barré syndrome, in Campylobacter jejuni isolates from humans, broiler chickens, and wild birds. Microorganisms. 2020;8(3):325. DOI: https://doi.org/10.3390/microorganisms8030325
- Hwang S., Ryu S., Jeon B. Roles of the superoxide dismutase SodB and the catalase KatA in the antibiotic resistance of Campylobacter jejuni. J. Antibiot. (Tokyo). 2013;66(6):351–3. DOI: https://doi.org/10.1038/ja.2013.20
Supplementary files