Antimicrobial resistance in foodborne Salmonella enterica isolates in the Republic of Belarus
- Authors: Kulikova N.G.1, Chernyshkov A.V.1, Mikhaylova Y.V.1, Zenkovich A.L.2, Dovnar D.A.2, Mareiko A.M.2, Bityumina L.A.1, Shelenkov A.A.1, Egorova A.E.1, Saenko S.S.1, Manzenyuk I.N.1
-
Affiliations:
- Central Research Institute for Epidemiology
- State Institution Republican Center for Hygiene, Epidemiology and Public Health
- Issue: Vol 100, No 2 (2023)
- Pages: 153-167
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/1301
- DOI: https://doi.org/10.36233/0372-9311-343
- EDN: https://elibrary.ru/mqqmxo
- ID: 1301
Cite item
Abstract
Introduction. Antimicrobial resistance is a global public health concern. Salmonella spp., which can be transmitted to humans through contaminated food, are among the most important foodborne pathogens worldwide.
Materials and methods. The antimicrobial resistance of 358 bacterial isolates collected from food and water in the Republic of Belarus (Belarus) in 2018–2021 was studied by analyzing phenotypic and genotypic characteristics of antibiotic bacterial resistance. MALDI-TOF mass spectrometry was used to classify and identify bacteria. Phenotypic antimicrobial susceptibility of bacteria was measured by the minimum inhibitory concentration method using a Sensititre automated bacteriological analyzer and the disk diffusion test for 45 antimicrobial agents. Antimicrobial resistance genes in multidrug-resistant Salmonella isolates were identified by whole-genome sequencing.
Results. The in vitro testing of phenotypic bacterial susceptibility showed high susceptibility to fluoroquinolones (97.2%), third-generation cephalosporins (93.9%), carbapenems (98.0%), ampicillin (81.8%), aminoglycosides (97.5%), tetracyclines (87.5%), chloramphenicol (93.8%), trimethoprim/sulfamethoxazole (co-trimoxazole) (95.3%) and colistin (85.2%). It was found that the antibiotic resistance mechanism in S. enterica was associated with the presence of genes blaTEM-1B (82%), blaTEM-1C (7.7%), blaSHV-12 (2.6%), blaDHA-1 (2.6%), blaCMY-2 (7.7%), qnrB2 (9.1%), qnrB4 (9.1%), qnrB5 (9.1%), qnrB19 (72.7%), aac(6’)-Ib-cr (9.1%), aac(6’)-Iaa (100%), aadA1 (13.2%), aadA2 (8.8%), tetB (74.3%), tetA (25.7%), tetM (2.9%), tetD (28.6%), mcr-9 (1.5%).
Conclusion. All the bacterial isolates were phenotypically susceptible to first-line antibiotics used in treatment of salmonellosis: fluoroquinolones and third-generation cephalosporins. The whole-genome sequencing of multidrug-resistant Salmonella isolates (19.0%) detected resistance genes for 9 groups of antibiotics: aminoglycosides (100%), beta-lactams (57.4%), fluoroquinolones (16.2%), tetracyclines (51.5%), macrolides (1.5%), phenicols (30.4%), trimethoprim (13.0%), sulfonamides (47.8%) and colistin (1.4%). Thus, epidemiological surveillance of the Salmonella spread through the food chain is of critical importance for the monitoring of antimicrobial resistance among foodborne Salmonella.
Full Text
Introduction
Among all foodborne pathogens, the leading role in bacterial invasion into the gastrointestinal tract belongs to different Salmonella enterica serovars [1]. Salmonella invasion in humans poses a great threat due to the ability of Salmonella to cause persistent infection and complications [2]. Having high environmental plasticity, S. enterica species can easily find ecological niches, adapt to different conditions, and remain viable in dry and frozen food products [1, 2]; they can also adapt to mass drug administration of antibiotics in public health and agriculture, thus contributing to increasing resistance to antimicrobial agents.
Drug resistance mechanisms of bacteria depend on different enzyme-mediated factors [3]. Considering that Salmonella spp. can act as a vector of transfer of resistance genes to other microorganisms, the studies of phenotypic and genotypic resistance profiles of Salmonella are highly important for monitoring of spread of antibiotic resistance.
Materials and methods
Collection of microorganisms
The study was performed using S. enterica cultures (n = 358) isolated in Belarus in 2018–2021. The isolation and primary identification of bacterial isolates were performed at the Republican Center of Hygiene, Epidemiology and Public Health (Minsk).
The sources of bacterial isolates were poultry (n = 113), meat (n = 52), fish (n = 1), dairy (n = 2), confectionery (n = 3), precooked and processed (n = 158) products, wastewater and washings collected from work surfaces (n = 29). The final species-level identification of bacterial isolates and assessment of their antimicrobial susceptibility were performed at the Rospotrebnadzor Reference Center for Monitoring the Residual Amount of Antibiotics and Antibiotic Resistance of Bacteria in Food Raw Materials and Food Products at the Rospotrebnadzor Central Research Institute of Epidemiology (Moscow).
Species-level identification and storage of bacterial isolates
All the studied bacterial isolates were identified to the genus level using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), the Microflex LT system and the MALDI Biotyper Compass v.4.1.80 software (Bruker Daltonics). The recommended score of ≥ 2.0 was used as a criterion for accurate species-level identification with MALDI-TOF mass-spectrometry. Serotyping of Salmonella was performed using the Salmonella sera agglutination test (PETSAL) in accordance with the Kauffmann–White classification scheme. Bacterial isolates were stored at –70ºC in Mueller–Hinton agar with 10% glycerol [4].
Assessment of susceptibility to antimicrobial agents
Antimicrobial susceptibility profiles of foodborne bacterial isolates obtained in 2018–2019 were evaluated using the disk diffusion method and the following antibiotics: ampicillin, cefotaxime, ceftazidime, meropenem, ciprofloxacin, levofloxacin, amikacin, gentamicin, chloramphenicol and co-trimoxazole. Clinical categories of antimicrobial susceptibility of bacterial isolates were identified with reference to the breakpoints for the minimum inhibitory concentration in accordance with the EUCAST guidelines (versions 8.0, 2018 and 9.0, 2019, respectively).
The antimicrobial susceptibility profiling of foodborne bacterial isolates collected in 2020–2021 was performed by microdilution in the Mueller–Hinton agar and measuring the minimum inhibitory concentration using a Sensititre semi-automated analyzer (TREK Diagnostics Systems). Bacterial inoculation was performed using 96-well RUGNF and GN4F microplates for gram-negative bacterial isolates. The test results for antimicrobial susceptibility of bacterial isolates from raw foods and food products were analyzed using the SWIN software in accordance with the CLSI interpretation guidelines (30th edition, 2020) and/or EUCAST (versions 10.0, 2020 and 11.0, 2021, respectively). E. coli ATCC25922 and E. coli ATCC35218 cultures were used for susceptibility assessment quality control.
Detection of genetic resistance determinants
The determinants of genetic resistance in multidrug-resistant Salmonella isolates were detected using whole-genome sequencing. The RIBO-prep reagent kit (Central Research Institute of Epidemiology) was used for DNA extraction. Samples for DNA sequencing were prepared using the Illumina Nextera DNA Library Prep Kit and Illumina Nextera Index Kit. The sequencing was performed with the Illumina HiSeq1500 system (Illumina), including Illumina HiSeq PE Rapid Cluster Kit v2 and Illumina HiSeq Rapid SBS Kit v2 reagent kits.
Bioinformatic analysis
Genome assemblies from short reads were obtained using SPAdes v. 3.12 [5] with default parameters. The assembly quality assessment, completeness evaluation and initial annotation were performed using the software that was described earlier [6]. The Resfinder 4.0 database [7], including default parameters, was used for in silico identification of antibiotic-resistance genes; typing of bacterial isolates was performed using the multilocus sequence typing (MLST) scheme and Pasteur MLST website1, as of 20/10/2021).
Statistical analysis of the results
Standard methods of descriptive statistics and Microsoft Office Excel 2010 were used for the statistical analysis of the study results. The statistical significance of differences in percentages of resistant cultures was assessed using Student's t-test and the threshold value α < 0.05.
Results
A total of 358 S. enterica isolates from raw foods and food products in Belarus were studied in 2018–2021. Most of the cultures were delivered for further studies to the Rospotrebnadzor Reference Center in 2018 (n = 121; 33.8 ± 0.29%); the smallest percentage of cultures were delivered in 2021 (n = 43; 12.0 ± 0.14%). In 2019 and 2020, the Reference Center received 104 (29.1 ± 0.27%) and 90 (25.1 ± 0.24%) bacterial isolates, respectively.
Most of the cultures were isolated from meat products (n = 52), poultry products (n = 113) and precooked products from processed pork and poultry (n = 158) (Table 1). The smallest number of Salmonella was isolated from confectionary, dairy and fish products. In addition to food products, Salmonella bacteria were isolated from drinking water, wastewater and washings collected from work surfaces, which were classified as other products (n = 29).
Table 1. Salmonella content level in food products
Source | Number of isolates | Number of isolates, % |
Cookery food | 158 | 44,1 ± 0,28 |
Poultry | 113 | 31,6 ± 0,24 |
Meat | 52 | 14,5 ± 0,14 |
Confectionery | 3 | 0,8 ± 0,06 |
Dairy | 2 | 0,6 ± 0,05 |
Seafood | 1 | 0,3 ± 0,04 |
Others | 29 | 8,1 ± 0,1 |
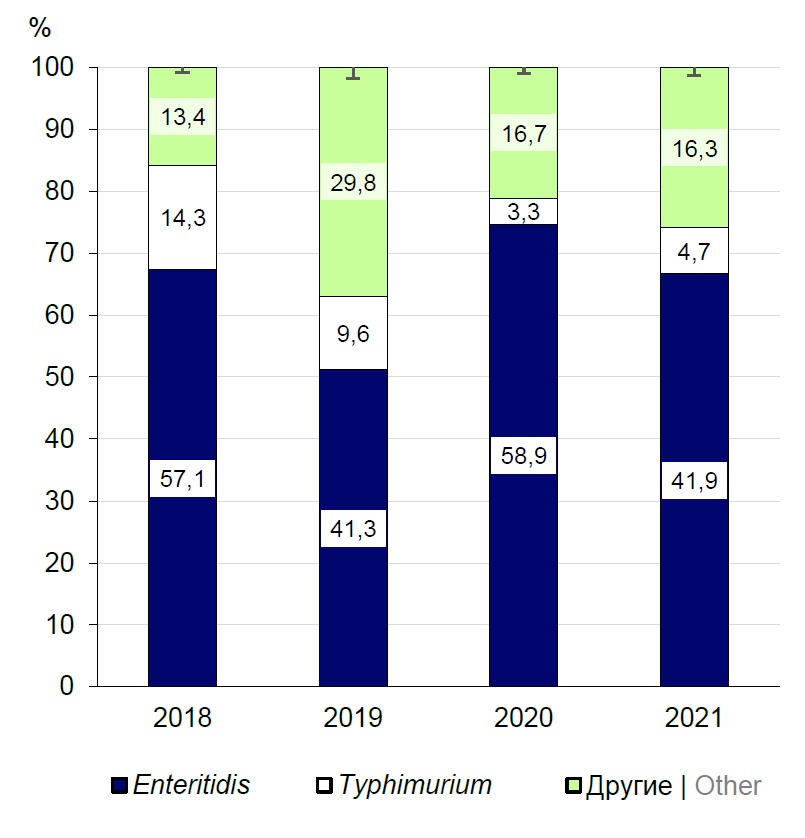
A total of 28 serotypes of S. enterica were identified during the studies. Serotype Enteritidis isolates accounted for the highest percentage (n = 182; 50.80 ± 0.20%): In 2018, they accounted for 57.10 ± 0.27% (n = 68), in 2019 — 41.30 ± 0.22% (n = 43), in 2020 — 58.9 ± 0.27% (n = 53), in 2021 – 41.90 ± 0.12% (n = 18) (Fig. 1). From 3.30 ± 0.12% (n = 3) in 2020 to 14.30 ± 0.56% (n = 17) of Salmonella bacteria in 2018 belonged to serotype Typhimurium (n = 61; 17.00 ± 0.15%). All the other serotypes were represented by the smallest percentage (from 0.30 ± 0.01% to 2.50 ± 0.06%), and therefore were assigned to the group of "other", which included from 13.40 ± 0.16% to 29.80 ± 0.25% of cultures. The cultures in this group belonged to serotypes Agona, Blegdam, Brandenburg, Bredeney, Chester, Derby, Dublin, Essen, Fyris, Give, Goettingen, Goma, Infantis, Jerusalem, Kapemba, Kottbus, London, Mbandaka, Munchen, Panama, Saintpaul, Sandiego, Tsevie, Virchow.
Fig. 1. Prevalence of foodborne S. enterica serotypes isolated in Belarus.
During the studies in 2018–2021, the analyzed data on phenotypic susceptibility of Salmonella isolates to 45 antibacterial agents showed high susceptibility of bacteria to these agents (76.90 ± 0.06%). Multidrug resistance (MDR) was found in 19.00 ± 0.05% (n = 68) of cultures.
The main medications for treatment of severe salmonellosis are fluoroquinolone antibiotics that have no cross-resistance with other classes of antibiotics due to their antimicrobial activity induced by inhibition of DNA gyrase or topoisomerase IV [8]. The analysis of phenotypic susceptibility of Salmonella bacteria isolated from food products and raw foods in Belarus demonstrated high susceptibility of bacteria to this group of antibiotics (from 88.40 ± 0.31% to 100%). However, the period of 2020–2021 demonstrated a tendency towards a gradual annual increase in the percentage of resistant S. enterica isolates: from 0% in 2018 and 2019 to 5.6 ± 0.1% and 11.60 ± 0.31% in 2020 and 2021, respectively (Fig. 2).
Fig. 2. Profile of phenotypic susceptibility S. enterica isolates to fluoroquinolones.
The analysis of phenotypic susceptibility of Salmonella showed the tendency towards decreasing activity of third-generation cephalosporins, though the percentage of phenotypically susceptible cultures remained high: from 100% in 2018 to 83.70 ± 0.14% in 2021 (Fig. 3). At the same time, the annual increase in phenotypically resistant Salmonella cultures was observed throughout the period of studies.
Fig. 3. Profile of phenotypic susceptibility of S. enterica isolates to third-generation cephalosporins.
During the period of studies, S. enterica isolates were phenotypically highly susceptible to such reserve antibiotics for salmonellosis treatment as ampicillin and carbapenems — imipenem and meropenem. Regarding ampicillin, there was a general trend towards a gradual increase in the percentage of resistant cultures from 14.9 ± 0.1% in 2018 to 23.30 ± 0.55% in 2021; as for carbapenems, the percentage of resistant cultures increased to 5.60 ± 0.11% in 2020 compared to 2018 and 2019; then, it slightly decreased to 4.70 ± 0.14% in 2021.
Antibiotics from the group of aminoglycosides are of primary clinical significance in treatment of nosocomial infections caused by aerobic gram-negative bacteria. The studies of phenotypic aminoglycoside susceptibility of S. enterica cultures isolated from food products in Belarus demonstrated high phenotypic susceptibility to aminoglycosides during the entire period of monitoring: from 95.30 ± 0.06% to 100.0%. However, in 2020–2021, the percentage of resistant cultures increased gradually to 3.30 ± 0.07% and 4.70 ± 0.15%, respectively (Fig. 4).
Fig. 4. Profile of phenotypic susceptibility of S. enterica isolates to aminoglycoside antibiotics.
In 2020 and 2021, the isolated Salmonella cultures were assessed for their susceptibility to colistin and tetracyclines as reserve antibiotics against multidrug resistant microorganisms. Colistin remains a drug of last resort, being used for treatment of life-threatening infections caused by carbapenem-resistant enterobacteria. Some countries and regions have already reported the existence of colistin-resistant bacteria causing infections, against which there are no effective antibiotics [9]. Our study revealed an upward trend in phenotypically colistin-resistant bacterial isolates, the percentage of which increased 2.3 times (from 10.10 ± 0.18% in 2020 to 23.30 ± 0.58% in 2021), and in tetracycline-resistant isolates, the percentage of which increased 7.2 times (from 3.9 ± 0.1% in 2020 to 27.90± 0.65% in 2021; Fig. 5). Broad-spectrum reserve antibiotics are represented by co-trimoxazole and chloramphenicol, which were characterized by a low percentage of resistant cultures throughout the monitoring period: from 4.8 ± 0.1 to 6.70 ± 0.13% and 3.80 ± 0.07% to 7.4 ± 0.12%, respectively.
Fig. 5. Changes in the percentage of S. enterica cultures phenotypically resistant to colistin and tetracycline in 2020–2021
The severity of Salmonella infection depends on multiple factors, including the presence of antimicrobial resistance determinants in bacteria [10]. In 2018–2021, in Belarus, a total of 68 (19.0 ± 0.2%) multidrug-resistant Salmonella isolates were identified and were further studied for genetic markers of resistance. The main mechanism of resistance to beta-lactam antibiotics in Salmonella spp. involves acquisition of bla genes, which encode enzymes capable of inactivating antibiotics [11]. Although the percentage of cultures phenotypically resistant to beta-lactam antibiotics was small, the genotypic profile of resistance of bacterial isolates showed a high percentage of producers of Class A and C beta-lactamases (n = 39; 57.4 ± 0.2%). Most of the bacterial isolates contained extended spectrum beta-lactamases (ESBLs) blaTEM-1B (n = 32; 82.10 ± 0.16%), blaTEM-1C (n = 3; 7.70 ± 0.26%), blaSHV-12 (n = 1; 2.60 ± 0.11%), blaDHA-1 (n = 1; 2.60 ± 0.11%); in addition, isolates of serotype Enteriditis were detected, which contained cephalosporinases blaCMY-2 (n = 3; 7.70 ± 0.26%; Table 2).
Table 2. Genotypic profile of beta-lactam antibiotic resistance of S. enterica isolates
Year | Isolate | Serotype | MLST | Resistance genes | ||||
blaTEM-1B | blaTEM-1C | blaCMY-2 | blaDHA-1 | blaSHV-12 | ||||
2018 | Crie F21 | Enteritidis | ST11 | + | – | – | – | – |
2018 | Crie F28 | Tythimurium | ST34 | + | – | – | – | – |
2018 | Crie F47 | Enteritidis | ST11 | – | + | – | – | – |
2018 | Crie F34 | Mendoza | ST490 | + | – | – | – | – |
2018 | Crie F50 | Tythimurium | ST34 | + | – | – | – | – |
2018 | Crie F40 | Tythimurium | ST34 | + | – | – | – | – |
2018 | Crie F51 | Enteritidis | ST11 | – | + | – | – | – |
2018 | Crie F297 | Enteritidis | ST11 | + | – | – | – | – |
2018 | Crie F46 | Tythimurium | ST9644 | + | – | – | – | – |
2018 | Crie F36 | Tythimurium | ST34 | + | – | – | – | – |
2018 | Crie F37 | Tythimurium | ST34 | + | – | – | – | – |
2018 | Crie F303 | Enteritidis | ST11 | + | – | – | – | – |
2019 | Crie F146 | Enteritidis | ST11 | + | – | – | – | – |
2019 | Crie F296 | Tythimurium | ST19 | + | – | – | – | – |
2019 | Crie F149 | Tythimurium | ST34 | + | – | – | – | – |
2019 | Crie F158 | Brandenburg | ST9644 | + | – | – | – | – |
2019 | Crie F298 | Mendoza | ST490 | + | – | – | – | – |
2019 | Crie F159 | Enteritidis | ST11 | – | + | – | – | – |
2019 | Crie F162 | Enteritidis | ST11 | + | – | – | – | – |
2019 | Crie F163 | Enteritidis | ST11 | + | – | – | – | – |
2019 | Crie F164 | Tythimurium | ST34 | + | – | – | – | – |
2019 | Crie F165 | Tythimurium | ST34 | + | – | – | – | – |
2019 | Crie F167 | Tythimurium | ST34 | + | – | – | – | – |
2019 | Crie F168 | Tythimurium | ST34 | + | – | – | – | – |
2019 | Crie F302 | Enteritidis | ST11 | – | – | + | – | – |
2019 | Crie F353 | Tythimurium | ST34 | + | – | – | – | – |
2020 | Crie F919 | Tythimurium | ST34 | + | – | – | – | – |
2020 | Crie F920 | Tythimurium | ST34 | + | – | – | – | – |
2020 | Crie F923 | Enteritidis | ST11 | – | – | + | – | – |
2020 | Crie F926 | Enteritidis | ST11 | – | – | + | – | – |
2021 | Crie F1149 | Tythimurium | ST34 | + | – | – | – | – |
2021 | Crie F1151 | Tythimurium | ST34 | – | – | – | + | + |
2021 | Crie F1153 | Tythimurium | ST34 | + | – | – | – | – |
2021 | Crie F1154 | Tythimurium | ST34 | + | – | – | – | – |
2021 | Crie F1155 | Tythimurium | ST34 | + | – | – | – | – |
2021 | Crie F1156 | Tythimurium | ST34 | + | – | – | – | – |
2021 | Crie F1157 | Tythimurium | ST34 | + | – | – | – | – |
2021 | Crie F1159 | Tythimurium | ST34 | + | – | – | – | – |
The analysis of the genotypic susceptibility profile of MDR Salmonella revealed the presence of fluoroquinolone resistance determinants in 11 isolates (16.20 ± 0.33%), which were encoded by qnrB2 (n = 1; 9.10 ± 0.69%), qnrB4 (n = 1; 9.10 ± 0.69%), qnrB5 (n = 1; 9.10 ± 0.69%), qnrB19 (n = 8; 72.70 ± 0.12%) genes and aminoglycoside acetyltransferase enzyme aac(6’)-Ib-cr (n = 1; 9.10 ± 0.69%), causing simultaneous inactivation of fluoroquinolones and aminoglycosides (Table 3).
Table 3. Genotypic profile of resistance of S. enterica to fluoroquinolones
Year | Isolate | Serotype | MLST | Resistance genes | ||||
qnrB2 | qnrB4 | qnrB5 | qnrB19 | aac(6')-Ib-cr | ||||
2018 | Crie F46 | Tythimurium | ST9644 | – | – | – | + | – |
2019 | Crie F298 | Mendoza | ST490 | + | – | – | – | – |
2019 | Crie F353 | Tythimurium | ST34 | – | – | – | + | – |
2020 | Crie F920 | Tythimurium | ST34 | – | – | – | + | – |
2020 | Crie F921 | Infantis | ST32 | – | – | – | + | – |
2020 | Crie F922 | Enteritidis | ST11 | – | – | – | + | – |
2020 | Crie F925 | Enteritidis | ST11 | – | – | – | + | – |
2020 | Crie F926 | Enteritidis | ST11 | – | – | – | + | – |
2021 | Crie F1149 | Tythimurium | ST34 | – | – | + | – | – |
2021 | Crie F1151 | Tythimurium | ST34 | – | + | – | – | + |
2021 | Crie F1159 | Tythimurium | ST34 | – | – | – | + | – |
Despite the high percentage of cultures phenotypically susceptible to aminoglycosides, the whole-genome sequencing showed that resistance determinants for this group of agents were present in all the studied MDR-cultures, including Salmonella, which demonstrated phenotypic susceptibility to aminoglycosides (n = 61; 89.70 ± 0.08%). The dominant resistance gene detected in all the bacterial isolates was aac(6’)-Iaa (n = 68; 100%). The main genetic markers of resistance to aminoglycosides, which were identified in our studies, were aadA1 genes (n = 9; 13.20 ± 0.28%) and aadA2 genes (n = 6; 8.8 ± 0.2%).
Mobilized colistin resistance (mcr) genes were found only in one MDR-culture — Crie F1151, which was phenotypically susceptible to colistin. No mcr genes were detected in cultures phenotypically resistant to colistin. Genetic determinants of tetracycline resistance were detected in 51.5 ± 0.2% (n = 35) of Salmonella bacteria (Table 4). The resistance mechanisms involved genes encoding efflux pumps of the cytoplasmic membrane: tetB (n = 26; 74.30 ± 0.65%), tetA (n = 9; 25.70 ± 0.67%), tetD (n = 10; 28.60 ± 0.71%); the studied cultures also had the tetracycline resistance gene tetM (n = 1; 2.90 ± 0.15%) protecting the target from tetracycline action.
Table 4. Genotypic profile of S. enterica resistance to chloramphenicol
Year | Isolate | Serotype | MLST | Resistance genes | |||
cmlA1 | floR | catA1 | catA2 | ||||
2018 | Crie F21 | Enteritidis | ST11 | + | – | – | – |
2018 | Crie F28 | Tythimurium | ST34 | – | + | – | – |
2018 | Crie F29 | Enteritidis | ST11 | – | – | + | – |
2018 | Crie F40 | Tythimurium | ST34 | – | + | – | |
2018 | Crie F299 | Enteritidis | ST11 | – | – | + | – |
2018 | Crie F36 | Tythimurium | ST34 | – | + | – | – |
2018 | Crie F37 | Tythimurium | ST34 | – | + | – | – |
2018 | Crie F303 | Enteritidis | ST11 | + | – | – | – |
2019 | Crie F146 | Enteritidis | ST11 | + | – | – | – |
2019 | Crie F149 | Tythimurium | ST34 | – | + | – | – |
2019 | Crie F298 | Mendoza | ST490 | – | + | – | – |
2019 | Crie F352 | Enteritidis | ST11 | – | – | + | – |
2019 | Crie F164 | Tythimurium | ST34 | – | + | – | – |
2019 | Crie F165 | Tythimurium | ST34 | – | + | – | – |
2019 | Crie F168 | Tythimurium | ST34 | – | + | – | – |
2019 | Crie F170 | Enteritidis | ST11 | – | – | + | – |
2019 | Crie F171 | Tythimurium | ST34 | – | – | + | – |
2020 | Crie F919 | Tythimurium | ST34 | + | + | – | – |
2020 | Crie F920 | Tythimurium | ST34 | – | + | – | – |
2021 | Crie F1151 | Tythimurium | ST34 | – | – | – | + |
The analysis of the results of our genotypic studies of MDR Salmonella cultures showed that 29.4 ± 0.51% (n = 20) of them had plasmid-mediated efflux pumps genes cmlA1 (n = 4; 20.00 ± 0.83%) and floR (n = 11; 55.00 ± 1.13%) responsible for resistance to phenicols as well as genes encoding chloramphenicol acetyltransferase enzyme – catA1 (n = 5; 25.00 ± 0.92%) and catA2 (n = 1; 5.00 ± 0.25%; Table 5).
Table 5. Genotypic profile of S. enterica resistance to tetracyclines
Year | Isolate | Serotype | MLST | Resistance genes | |||
tetB | tetA | tetM | tetD | ||||
2018 | Crie F28 | Tythimurium | ST34 | + | – | – | – |
2018 | Crie F29 | Enteritidis | ST11 | – | + | – | – |
2018 | Crie F34 | Mendoza | ST490 | + | – | – | – |
2018 | Crie F50 | Tythimurium | ST34 | + | – | – | – |
2018 | Crie F40 | Tythimurium | ST34 | + | – | – | – |
2019 | Crie F296 | Tythimurium | ST19 | – | + | – | – |
2019 | Crie F147 | Infantis | ST32 | – | + | – | – |
2019 | Crie F149 | Tythimurium | ST34 | + | – | – | – |
2019 | Crie F158 | Brandenburg | ST9644 | + | – | – | – |
2019 | Crie F298 | Mendoza | ST490 | + | – | – | – |
2019 | Crie F352 | Enteritidis | ST11 | – | + | – | – |
2019 | Crie F164 | Tythimurium | ST34 | + | – | – | – |
2019 | Crie F165 | Tythimurium | ST34 | + | – | – | – |
2019 | Crie F167 | Tythimurium | ST34 | + | – | – | – |
2019 | Crie F168 | Tythimurium | ST34 | + | – | – | – |
2019 | Crie F170 | Enteritidis | ST11 | – | + | – | – |
2019 | Crie F171 | Enteritidis | ST11 | – | + | – | – |
2019 | Crie F353 | Tythimurium | ST34 | + | – | – | – |
2018 | Crie F46 | Tythimurium | ST9644 | + | – | – | – |
2018 | Crie F299 | Enteritidis | ST11 | – | + | – | – |
2018 | Crie F36 | Tythimurium | ST34 | + | – | – | – |
2018 | Crie F37 | Tythimurium | ST34 | + | – | – | – |
2020 | Crie F919 | Tythimurium | ST34 | + | – | + | – |
2020 | Crie F920 | Tythimurium | ST34 | + | – | – | – |
2020 | Crie F921 | Infantis | ST32 | – | + | – | – |
2021 | Crie F1148 | Virchow | ST8662 | – | + | – | – |
2021 | Crie F1149 | Tythimurium | ST34 | + | – | – | – |
2021 | Crie F1150 | Tythimurium | ST34 | + | – | – | – |
2021 | Crie F1151 | Tythimurium | ST34 | + | – | – | + |
2021 | Crie F1153 | Tythimurium | ST34 | + | – | – | – |
2021 | Crie F1154 | Tythimurium | ST34 | + | – | – | – |
2021 | Crie F1155 | Tythimurium | ST34 | + | – | – | – |
2021 | Crie F1156 | Tythimurium | ST34 | + | – | – | – |
2021 | Crie F1157 | Tythimurium | ST34 | + | – | – | – |
2021 | Crie F1159 | Tythimurium | ST34 | + | – | – | – |
Co-trimoxazole resistance determinants were detected in 50.0 ± 0.2% (n = 34) of MDR cultures with the genotypic profile of resistance being represented by dihydrofolate reductase genes dfrA1 (n = 2.00 ± 0.18; 5.9%), dfrA8 (n = 2.00 ± 0.18; 5.9%), dfrA12 (n = 3.00 ± 0.27; 8.8%) and dfrA14 (n = 2.00 ± 0.18; 5.9%) and by genes (n = 33; 48.50 ± 0.59%) expressing dihydropteroate synthases resistant to sulfonamides – sul1 (n = 6; 18.2 ± 0.59%), sul2 (n = 23; 69.7 ± 0.74%) and sul3 (n = 4; 12.1 ± 0.36%; Table 6).
Table 6. Genotypic profile of S. enterica resistance to co-trimoxazole
Year | Isolate | Serotype | ST | Resistance genes | ||||||
sul3 | sul2 | sul1 | dfrA12 | dfrA8 | dfrA14 | dfrA1 | ||||
2018 | Crie F21 | Enteritidis | ST11 | + | – | – | – | – | – | – |
2018 | Crie F28 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2018 | Crie F34 | Mendoza | ST490 | – | – | + | + | – | – | – |
2018 | Crie F50 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2018 | Crie F40 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2018 | Crie F46 | Tythimurium | ST9644 | – | + | – | – | – | – | – |
2018 | Crie F36 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2018 | Crie F37 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2018 | Crie F303 | Enteritidis | ST11 | + | – | – | – | + | – | – |
2019 | Crie F146 | Enteritidis | ST11 | + | – | – | – | – | – | – |
2019 | Crie F296 | Tythimurium | ST19 | + | – | – | – | – | – | – |
2019 | Crie F147 | Infantis | ST32 | – | – | + | – | – | – | – |
2019 | Crie F149 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2019 | Crie F158 | Brandenburg | ST9644 | – | + | – | – | – | – | – |
2019 | Crie F298 | Mendoza | ST490 | – | – | + | + | – | – | – |
2019 | Crie F164 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2019 | Crie F165 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2019 | Crie F166 | Enteritidis | ST11 | – | – | – | – | + | – | – |
2019 | Crie F167 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2019 | Crie F168 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2019 | Crie F353 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2020 | Crie F919 | Tythimurium | ST34 | – | + | – | + | – | – | – |
2020 | Crie F920 | Tythimurium | ST34 | – | + | – | – | – | + | – |
2020 | Crie F921 | Infantis | ST32 | – | – | + | – | – | + | – |
2021 | Crie F1148 | Virchow | 8662 | – | – | + | – | – | – | + |
2021 | Crie F1149 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2021 | Crie F1150 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2021 | Crie F1151 | Tythimurium | ST34 | – | – | + | – | – | – | + |
2021 | Crie F1153 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2021 | Crie F1154 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2021 | Crie F1155 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2021 | Crie F1156 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2021 | Crie F1157 | Tythimurium | ST34 | – | + | – | – | – | – | – |
2021 | Crie F1159 | Tythimurium | ST34 | – | + | – | – | – | – | – |
Note that despite the presence of antibiotic resistance determinants, 10 cultures were susceptible to all the studied antibiotics – Crie F146, Crie F149, Crie F158, Crie F159, Crie F162, Crie F163, Crie F164, Crie F165, Crie F167 and Crie F168. In addition, in our study, we did not detect carbapenemases of class A (KPC) and class B (GIM, VIM, IMP, NDM, SPM and FOX).
Almost all Salmonella cultures producing ESBLs or AmpC (n = 20; 51.30 ± 0.27%) were characterized by complete phenotypic susceptibility to other, non-beta-lactam antibiotics, including fluoroquinolones; 46.20 ± 0.27% (n = 18) of bacterial isolates were resistant to 1–2 non-lactam antibiotics.
The in silico multilocus sequence typing analysis revealed 10 different sequence types of MDR S. enterica isolates: serovar Enteritidis ST11 (n = 31; 47.0 ± 0.57%), serovar Typhimurium ST34 and ST19 (n = 21; 33.30 ± 0.51% and n = 3; 4.50 ± 0.10%, respectively), serovar Infantis ST32 (n = 4; 6.10 ± 0.13%), serovar Mendoza ST490 (n = 3; 4.50 ± 0.10%), serovar Bredeney ST897 (n = 1; 1.50 ± 0.03%), serovar Virchow ST8662 (n = 1; 1.50 ± 0.03%), serovar London ST1992 (n = 1; 1.00 ± 0.03%), serovar Stanleyville ST1986 (n = 1; 1.00 ± 0.03%). The new sequence type, ST9644, was identified in Crie F46 and Crie F158 Salmonella cultures. Dominant sequence types ST11, ST34, ST32, ST490 and ST19 were associated with multidrug resistance of the cultures that contained resistance determinants to 7 classes of antibiotics (Tables 2–6), while sequence types ST897, ST1992 and ST1986 contained genes responsible for resistance only to aminoglycosides.
The Crie F1151 culture was isolated from processed and precooked food products; it taxonomically belonged to Salmonella enterica serotype Typhimurium. The culture was characterized by phenotypic multidrug resistance to penicillins, cephalosporins, aztreonam, fluoroquinolones, aminoglycosides, trimethoprim/sulfamethoxazole and tetracyclines encoded by the respective resistance determinants: aac(6')-IIc, aac(6')-Iaa, aadA1, aph(3')-Ia, blaDHA-1, blaSHV-12, blaTEM-1B, aac(6')-Ib-cr, catA2, qnrB4, sul1, dfrA1 and tetB, tetD, respectively. The genotypic resistance profile of S. typhimurium Crie F1151 culture also included determinants responsible for resistance to ereA macrolides. This Salmonella isolate was the only one that had the mcr-9 determinant, despite the phenotypic susceptibility to colistin, as well as the aac(6')-Ib-cr determinant responsible for concurrent resistance to fluoroquinolones and aminoglycosides.
Discussion
Salmonellosis is a frequent gastrointestinal infection in humans and a major cause of foodborne disease outbreaks worldwide. In 2019, 87,923 confirmed cases of salmonellosis were reported in the European Union (EU); 57,702 cases were reported in 2020, being the lowest number reported since 2007 due to the exit of the United Kingdom from EU and the COVID-19 pandemic [9].
We studied Salmonella enterica cultures isolated from different food products in Belarus in 2018–2021 for the further assessment of their susceptibility to antibiotics. Our findings showed that pork and poultry products, including processed products, were the most frequent sources of Salmonella. Speaking about poultry products, we should note that the prevalence of resistant Salmonella increased significantly during the period of studies — from 19.8% in 2018 to 65.1% in 2021. The prevalence of Salmonella primarily in pork, chicken and turkey products was comparable with the rates reported by the United States, Egypt and Columbia [12–14]. Throughout the period of studies, the prevailing serotypes were Enteritidis (50.8%) and Typhimurium (9.0%). The dominance of these serotypes in meat products was also reported by researchers from India and Saudi Arabia, where serotypes Enteritidis and Typhimurium accounted for more than 95% of isolates [3, 11]. According to the European Union One Health Zoonoses Report (2020), serotype Enteritidis also dominated in the European Region [15].
The analysis of phenotypic resistance of cultures isolated in Belarus revealed high susceptibility to antibiotics of the fluoroquinolone group, ranging from 88.4% to 100%. During the monitoring period, the increase in resistant cultures reached 11.6%. In the United States, during 2018–2021, the Centers for Disease Control and Prevention2 reported an 8.5% increase in Salmonella resistance to ciprofloxacin. High activity of fluoroquinolones was also found for Salmonella isolated from pork in Thailand: 76% of the studied cultures were susceptible. S. enteritidis, the most common type of Salmonella in humans, demonstrated the tendency to increased resistance to antibiotics of the fluoroquinolone group. According to the data from the European Centre for Disease Prevention and Control, in animals the resistance of S. enteritidis to these antibiotics ranged from moderate to high [16].
The phenotypic resistance of Salmonella isolated in Belarus to third-generation cephalosporins was not high, reaching 16.3%. During 2018–2021, the percentage of cultures resistant third-generation cephalosporins increased 5.6 times: from 2.9% in 2019 to 16.3% in 2021. According to the data from the Centers for Disease Control and Prevention, in the United States3 the percentage of cultures resistant to cephalosporins in the specified period was not large: 2.3% in 2018, 1.7% in 2019 and it remained stable at the level of 2% in 2020 and 2021. The report published by the European Centre for Disease Prevention and Control and the European Food Safety Authority states that in 2019, the percentage of cefotaxime-resistant and ceftazidime-resistant cultures in the European Region remained at low levels — 1.8% and 1.2%, respectively [17].
The increasing prevalence of MDR Salmonella poses a significant threat to public health, as it leads to longer hospital stays, longer duration of disease and higher fatality rates compared to susceptible Salmonella isolates [17, 18]. In 2021, in its report, the European Centre for Disease Prevention and Control pointed out that the percentage of MDR S. enterica isolates from pork and its products increased dramatically to 56.5% [17]. The World Health Organization estimates that of the 100,000 cases of salmonellosis each year, a large number are caused by MDR S. enterica [19], with the majority acquired through the consumption of contaminated food of animal origin, particularly beef, pork, and poultry products [20, 21]. Among the studied Salmonella isolates from Belarus, 19% of isolates demonstrated the MDR profile with resistance to 3 and more classes of antimicrobial agents, thus showing the consistency with studies by Egyptian researchers [22]. In 2008–2017, in the United States, resistance to 3 and more agents was detected in 28.0% of the bacteria isolated from poultry products [23]. Chinese researchers found that MDR was demonstrated by 95.33% of Salmonella isolated from pork [24]; Thai researchers also reported multidrug resistance of 23.2% of Salmonella isolated from duck meat [25].
The in silico multilocus sequence typing analysis of MDR Salmonella isolated in Belarus identified 5 sequence types of S. enterica, which were associated with multidrug resistance of Salmonella cultures. The dominant sequence types were represented by ST11 of serovar Enteritidis (47.0 ± 0.57%), ST34 (33.3 ± 0.51%) and ST19 (4.5 ± 0.10%) of serovar Typhimurium. These sequence types were also common in China and Iraq, where ST19 prevailed among serovar Typhimurium Salmonella [26–28]. In the European Region, ST11 was a prevailing sequence type of serovar Enteritidis, while in Russia, serotype Infantis ST32 was dominant [29].
The genotypic studies of cultures isolated in Belarus identified 5 genes responsible for resistance to beta-lactam antibiotics: blaTEM-1B, blaTEM-1C, blaDHA-1, blaSHV-12, as well as cephalosporinase blaCMY-2 genes. The annual increase in the percentage of phenotypically resistant cultures is most likely associated with the activation of resistance genes encoding ESBLs, as the studies showed that 56.5% of all the tested MDR isolates produced ESBLs. The blaSHV gene was identified mainly in representatives of the family Enterobacteriaceae, which were isolated from different ecosystems: humans, animals, and environment [30, 31]. Likely originated from a chromosomal penicillinase of Klebsiella pneumoniae, SHV beta-lactamases currently encompass a large number of allelic variants including ESBLs, non-ESBLs and several non-classified variants; therefore, their significance was emphasized in our studies [32].
The phenotypic characteristics of most MDR cultures (n = 59; 85.5%) correlated with the molecular mechanism of resistance and resulted from the spectrum of enzyme activity of beta-lactamases. The obtained data demonstrating the complete correlation between the phenotypic and genotypic characteristics of cultures from Belarus regarding their resistance to third-generation cephalosporins confirmed the diagnostic significance of such indicator substances as ceftazidime, ceftriaxone, cefoperazone and cefotaxime. During their three-year monitoring of Salmonella isolated from different categories of food products, the European laboratories also proved the diagnostic value of ceftriaxone, ceftazidime and cefotaxime substances for detection of cephalosporinases [9]. A sharp increase in the percentage of cultures phenotypically resistant to tetracycline — from 3.9% to 23.3%, along with a high percentage of cultures (52.2%) containing resistance determinants, can be indicative of excessive use of tetracycline antibiotics in agriculture, resulting in accumulation and transfer of antibiotic-resistance genes among pathogenic bacteria. The presence of antibiotic-resistance determinants in susceptible cultures as well as the presence of ereA and mcr-9 genes in S. typhimurium Crie F-1151 can serve as proof of Salmonella’s ability to act as a vector for transfer of antibiotic-resistance genes to other microorganisms. According to the published data, mcr-9 determinants, together with mcr-1 determinants, are considered the most common in the world [33]. Based on the data from the National Database of Antibiotic Resistant Organisms, the United States takes the lead by the number of mcr-9-positive isolates. In Europe, Russia and China, mcr-9 and mcr-1 determinants prevail [33].
The findings are certainly alarming, as all the studied bacterial isolates were received from samples of food products intended for human consumption. Although the risk of foodborne diseases can be reduced by heat treatment of food products, antibiotic-resistance genes may persist and, when entering the host, can be transferred to intestinal microbiocenosis, passing the resistance to other microorganisms [34]. Thus, our findings fall in line with the latest recommendations of the European Food Safety Authority that emphasized the significance of studying phenotypic and genotypic characteristics of foodborne bacterial isolates for monitoring and surveillance of antibiotic resistance, especially, for implementing the One Health approach that recognizes that the health of people is closely connected with the health of animals and the environment.
Conclusion
The irrational use of antibiotics in human and veterinary medicine has greatly contributed to the emergence and spread of resistant isolates of non-typhoid Salmonella.
The findings of our studies regarding the slowly growing phenotypic resistance to first-line antibiotics (cephalosporins and fluoroquinolones) and the presence of plasmid-mediated resistance determinants imply the possibility of a seriously limited choice of effective antimicrobial agents in future. Therefore, monitoring of antimicrobial resistance phenotypes and genotypes as well as transmission routes of Salmonella enterica cultures through the food chain is critically important.
The existing large diversity of resistance determinants and high phenotypic susceptibility of isolates lead to assumption that sources of bacterial isolates could be affected by antibiotics and/or could acquire resistance determinants from other microorganisms.
The conducted studies demonstrate the need for the further monitoring of prevalence of antibiotic resistant bacteria of food origin in Belarus, especially in the context of the One Health approach.
1 URL: https://bigsdb.pasteur.fr/
2 NARMS Now: Human Data.
URL: https://wwwn.cdc.gov/narmsnow/
3 Ibid.
About the authors
Nina G. Kulikova
Central Research Institute for Epidemiology
Author for correspondence.
Email: kulikova_ng@cmd.su
ORCID iD: 0000-0002-1716-6969
Cand. Sci. (Biol.), senior researcher, Laboratory of human clinical microbiology and microbial ecology
Russian Federation, MoscowAlexey V. Chernyshkov
Central Research Institute for Epidemiology
Email: chernyshkov@cmd.su
ORCID iD: 0000-0002-7049-4195
Cand. Sci. (Med.), senior researcher, Laboratory of human clinical microbiology and microbial ecology
Russian Federation, MoscowYuliya V. Mikhaylova
Central Research Institute for Epidemiology
Email: mihailova@cmd.su
ORCID iD: 0000-0002-5646-538X
Cand. Sci. (Biol.), Head, Laboratory for molecular mechanisms of antibiotic resistance
Russian Federation, MoscowAlexander L. Zenkovich
State Institution Republican Center for Hygiene, Epidemiology and Public Health
Email: kulikova_ng@cmd.su
deputy chief physician of the laboratory case
Belarus, MinskDaria A. Dovnar
State Institution Republican Center for Hygiene, Epidemiology and Public Health
Email: kulikova_ng@cmd.su
doctor, Microbiological reference laboratory
Belarus, MinskAla M. Mareiko
State Institution Republican Center for Hygiene, Epidemiology and Public Health
Email: kulikova_ng@cmd.su
doctor, Microbiological reference laboratory
Belarus, MinskLyutsiya A. Bityumina
Central Research Institute for Epidemiology
Email: bitumina@cmd.su
ORCID iD: 0000-0002-5378-0827
bacteriologist, Laboratory of human clinical microbiology and microbial ecology
Russian Federation, MoscowAndrey A. Shelenkov
Central Research Institute for Epidemiology
Email: shelenkov@cmd.su
ORCID iD: 0000-0002-7409-077X
Cand. Sci. (Appl. Math.), senior researcher, Laboratory of molecular mechanisms of antibiotic resistance
Russian Federation, MoscowAnna E. Egorova
Central Research Institute for Epidemiology
Email: a.egorova@cmd.su
ORCID iD: 0000-0003-0486-1353
researcher, Laboratory of molecular mechanisms of antibiotic resistance
Russian Federation, MoscowStepan S. Saenko
Central Research Institute for Epidemiology
Email: saenko@cmd.su
ORCID iD: 0000-0002-4925-1308
junior researcher, Laboratory of molecular mechanisms of antibiotic resistance
Russian Federation, MoscowIgor N. Manzenyuk
Central Research Institute for Epidemiology
Email: manzeniuk@cmd.su
ORCID iD: 0000-0002-1146-1430
Cand. Sci. (Med.), assistant director for research
Russian Federation, MoscowReferences
- Pavlova A.S., Bocharova Yu.A., Kuleshov K.V., et al. Molecular determinants of antibiotic resistance in Salmonella enterica antibiotic resistance. Journal of microbiology, epidemiology and immunobiology. 2021;98(6):721–30. DOI: https://doi.org/10.36233/0372-9311-140. EDN: https://elibrary.ru/zqrygu
- Reshetneva I.T., Per'yanova O.V., Dmitrieva G.M., Ostapova T.S. Antibiotic resistance of Salmonella spp. isolated in the territory of the Krasnoyarsk region. Hygiene and Sanitation. 2015;94(2):35–8. EDN: https://elibrary.ru/tphjlx
- Al-Ansari M.M., Aljubali M.M., Somily A.M., et al. Isolation and molecular characterization of multidrug-resistant Salmonella enterica serovars. J. Infect. Public Health. 2021;14(12):1767–76. DOI: https://doi.org/10.1016/j.jiph.2021.10.011
- Pokhilenko V.D., Baranov A.M., Detushev K.V. Methods of long-term storage of collection cultures of microorganisms and development trends. Izvestiya vysshikh uchebnykh zavedeniy. Povolzhskiy region. Meditsinskie nauki. 2009;(4):99–121. EDN: https://elibrary.ru/lakpfx
- Bankevich A., Nurk S., Antipov D., et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–77. DOI: https://doi.org/10.1089/cmb.2012.0021
- Shelenkov A., Mikhaylova Y., Yanushevich Y., et al. Molecular typing, characterization of antimicrobial resistance, virulence profiling and analysis of whole-genome sequence of clinical Klebsiella pneumoniae isolates. Antibiotics (Basel). 2020;9(5):261. DOI: https://doi.org/10.3390/antibiotics9050261
- Zankari E., Hasman H., Cosentino S., et al. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67(11):2640–4. DOI: https://doi.org/10.1093/jac/dks261
- Yakovlev V.P. Antimicrobial preparations of fluoroquinolones group. Consilium Medicum. 2006;8(1):35–41. EDN: https://elibrary.ru/rglbxl
- Gambino D., Gargano V., Butera G., et al. Food is reservoir of MDR Salmonella: prevalence of ESBLs profiles and resistance genes in strains isolated from food. Microorganisms. 2022;10(4):780. DOI: https://doi.org/10.3390/microorganisms10040780
- Ngoi S.T., Teh C.S., Chai L.C., Thong K.L. Overview of molecular typing tools for the characterization of Salmonella enterica in Malaysia. Biomed. Environ. Sci. 2015;28(10):751–64. DOI: https://doi.org/10.3967/bes2015.105
- Kumar Y., Singh V., Kumar G., et al. Serovar diversity of Salmonella among poultry. Indian J. Med. Res. 2019;150(1):92–5. DOI: https://doi.org/10.4103/ijmr.IJMR_1798_17
- Mamber S.W., Mohr T., Barlow K., et al. Occurrence of Salmonella in ready-to-eat meat and poultry product samples from U.S. Department of Agriculture-Regulated Producing Establishments. II. Salmonella in ready-to-eat pork barbecue products, from 2005 to 2012. J. Food Prot. 2018;81(10):1737–42. DOI: https://doi.org/10.4315/0362-028X.JFP-18-026
- Rodríguez R., Fandiño C., Donado P., et al. Characterization of Salmonella from commercial egg-laying hen farms in a central region of Colombia. Avian Dis. 2015;59(1):57–63. DOI: https://doi.org/10.1637/10873-052714-reg
- Elkenany R., Elsayed M.M., Zakaria A.I., et al. Antimicrobial resistance profiles and virulence genotyping of Salmonella enterica serovars recovered from broiler chickens and chicken carcasses in Egypt. BMC Vet. Res. 2019;15(1):124. DOI: https://doi.org/10.1186/s12917-019-1867-z
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2020 Zoonoses Report. EFSA J. 2021;19(12):e06971. DOI: https://doi.org/10.2903/j.efsa.2021.6971
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019-2020. EFSA J. 2022;20(3):e07209. DOI: https://doi.org/10.2903/j.efsa.2022.7209
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021;19(4):e06490. DOI: https://doi.org/10.2903/j.efsa.2021.6490
- Proietti P.C., Stefanetti V., Musa L., et al. Genetic profiles and antimicrobial resistance patterns of Salmonella infantis strains isolated in Italy in the food chain of broiler meat production. Antibiotics (Basel). 2020;9(11):814. DOI: https://doi.org/10.3390/antibiotics9110814
- WHO. Antimicrobial Resistance: Global Report on Surveillance. Geneva; 2014.
- Castro-Vargas R.E., Herrera-Sánchez M.P., Rodríguez-Hernández R., Rondón-Barragán I.S. Antibiotic resistance in Salmonella spp. isolated from poultry: a global overview. Vet. World. 2020;13(10):2070–84. DOI: https://doi.org/10.14202/vetworld.2020.2070-2084
- Campos J., Mourão J., Peixe L., Antunes P. Non-typhoidal Salmonella in the pig production chain: a comprehensive analysis of its impact on human health. Pathogens. 2019;8(1):19. DOI: https://doi.org/10.3390/pathogens8010019
- Lamas A., Fernandez-No I.C., Miranda J.M., et al. Prevalence, molecular characterization and antimicrobial resistance of Salmonella serovars isolated from northwestern Spanish broiler flocks (2011–2015). Poult. Sci. 2016;95(9):2097–105. DOI: https://doi.org/10.3382/ps/pew150
- Yin X., M'ikanatha N.M., Nyirabahizi E., et al Antimicrobial resistance in non-Typhoidal Salmonella from retail poultry meat by antibiotic usage-related production claims – United States, 2008–2017. Int. J. Food Microbiol. 2021;342:109044. DOI: https://doi.org/10.1016/j.ijfoodmicro.2021.109044
- Xu Z., Chen X., Tan W., et al. Prevalence and antimicrobial resistance of Salmonella and Staphylococcus aureus in fattening pigs in Hubei Province, China. Microb. Drug Resist. 2021;27(11): 1594–602. DOI: https://doi.org/10.1089/mdr.2020.0585
- Sinwat N., Witoonsatian K., Chumsing S., et al. Antimicrobial resistance phenotypes and genotypes of Salmonella spp. isolated from commercial duck meat production in Thailand and their minimal inhibitory concentration of disinfectants. Microb. Drug Resist. 2021;27(12):1733–41. DOI: https://doi.org/10.1089/mdr.2020.0230
- Yang J., Zhang Z., Zhou X., et al. Prevalence and characterization of antimicrobial resistance in Salmonella enterica isolates from retail foods in Shanghai, China. Foodborne Pathog. Dis. 2020;17(1):35–43. DOI: https://doi.org/10.1089/fpd.2019.2671
- Zhao X., Hu M., Zhang Q., et al. Characterization of integrons and antimicrobial resistance in Salmonella from broilers in Shandong, China. Poult. Sci. 2020;99(12):7046–54. DOI: https://doi.org/10.1016/j.psj.2020.09.071
- Harb A., Habib I., Mezal E.H., et al. Occurrence, antimicrobial resistance and whole-genome sequencing analysis of Salmonella isolates from chicken carcasses imported into Iraq from four different countries. Int. J. Food Microbiol. 2018; 284:84–90. DOI: https://doi.org/10.1016/j.ijfoodmicro.2018.07.007
- Egorova A., Mikhaylova Y., Saenko S., et al. Comparative whole-genome analysis of Russian foodborne multidrug-resistant Salmonella infantis isolates. Microorganisms. 2021;10(1):89. DOI: https://doi.org/10.3390/microorganisms10010089
- Alcalá L., Alonso C.A., Simón C., et al. Wild birds, frequent carriers of extended-spectrum β-lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 types. Microb. Ecol. 2016; 72(4):861–9. DOI: https://doi.org/10.1007/s00248-015-0718-0
- Liakopoulos A., Mevius D., Ceccarelli D. A review of SHV extended-spectrum β-lactamases: neglected yet ubiquitous. Front. Microbiol. 2016;7:1374. DOI: https://doi.org/10.3389/fmicb.2016.01374
- Bush K. Past and present perspectives on β-lactamases. Antimicrob. Agents Chemother. 2018;62(10):e01076–18. DOI: https://doi.org/10.1128/AAC.01076-18
- Ling Z., Yin W., Shen Z., et al. Epidemiology of mobile colistin resistance genes MCR-1 to MCR-9. J. Antimicrob. Chemother. 2020;75(11):3087–95. DOI: https://doi.org/10.1093/jac/dkaa205
- Groussin M., Poyet M., Sistiaga A., et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell. 2021;184(8):2053–67.e18. DOI: https://doi.org/10.1016/j.cell.2021.02.052
Supplementary files