Type 4 secretion system in Clostridioides difficile: Structural features and its role as a pathogenicity factor
- Authors: Sorokina J.V.1, Belyi Y.F.1
-
Affiliations:
- Gamaleya Research Centre for Epidemiology and Microbiology
- Issue: Vol 100, No 4 (2023)
- Pages: 345-353
- Section: REVIEWS
- URL: https://microbiol.crie.ru/jour/article/view/7271
- DOI: https://doi.org/10.36233/0372-9311-386
- EDN: https://elibrary.ru/rpsjli
- ID: 7271
Cite item
Abstract
Clostridioides difficile is a gram-positive microorganism causing damage to the human intestinal wall, clinically manifesting as antibiotic-associated diarrhea and pseudomembranous colitis. C. difficile infection remains a serious problem; the increasing frequency of nosocomial outbreaks and the emergence of community-acquired forms heighten the need for new prevention and treatment methods. The pathogenesis of C. difficile infection is associated with the toxins produced by bacteria and a large group of proteins promoting the replication of the pathogen in host tissues and its spread in the human population. Recent studies show that mobile genetic elements play a key role in the high virulence of C. difficile. Type 4 secretion systems (T4SS) are significant components of these elements; their impressive diversity among gram-positive microorganisms in general and in C. difficile, in particular, implies their high evolutionary and, consequently, medical significance. Further studies of the T4SS composition and structure will provide a deeper insight into mechanisms underlying the development of respective infections and will help outline pathogenically grounded approaches to prevention and treatment of diseases caused by C. difficile. On the other hand, the key components of the secretion machinery of the pathogen can be used in bioinformatic analysis and for searching new adaptive clusters in the genome of highly virulent strains.
Full Text
Introduction
Clostridioides difficile is a gram-positive, motile, spore-forming microorganism causing intestinal lesions in humans – antibiotic-associated diarrhea and colitis of varying severity [1–3]. The pathogenicity of C. difficile is mediated by the ability of the pathogen to produce at least one of the two glucosylating toxins – TcdA and TcdB [4, 5] – as well as the binary toxin (CDT), though the role of the latter is still not clearly understood [6–8]. The severity of the disease, the risk of developing complications and the transition into chronic forms, as well as the emergence of new endemic strains are often associated with additional factors responsible for adhesive functions [9], spore formation [10], biofilm formation [11], cell wall modification [12, 13], and transcription [14–16]. In addition, quorum-sensing proteins play an important role in the pathogenesis of C. difficile infection [17, 18], regulating, among other things, toxin production levels [19], while a large group of antibiotic resistance genes facilitates the unhindered development of infection during the antimicrobial treatment [20–23].
Nucleotide sequences presumably encoding components of the type 4 secretion system (T4SS) have been found in the genomes of C. difficile strains relatively recently [24]. This secretion machinery plays a key role in the pathogenesis of infections caused by gram-negative bacteria Agrobacterium tumefaciens [25], Legionella pneumophila [26], Helicobacter pylori, and others [27]. Meanwhile, the T4SS association with the pathogenesis of diseases caused by gram-positive pathogens has only recently gained attention of microbiologists [28–31]. It has become increasingly clear that exploration of the structure and funtction of this secretion machinery is important not only for deciphering infectious processes, but also for developing treatment and prevention tools [28, 30]. In our review, we tried to provide insights into distinctive features of the C. difficile T4SS class C organization to outline avenues and prospects of its further research.
Type 4 secretion system in gram-positive and gram-negative microorganisms
T4SS is a multicomponent transmembrane protein structure, which participates in the delivery of toxic effectors to the target cell [25], in the horizontal transfer of mobile genetic elements (MGEs) between microorganisms [23] and in the DNA exchange with the environment [27]. By their structure, the type 4 secretion systems are divided into three classes: A, B, and C (T4SS-A, T4SS-B, T4SS-C, respectively) [32, 33]. The latter is found only in gram-positive bacteria and is an integral part of the conjugative DNA transfer elements: plasmids, integrative and conjugative elements, and pathogenicity islands [33–35].
T4SS-A of the gram-negative phytopathogen A. tumefaciens is a prototype for the type 4 secretion machinery (Fig. 1, a). It is known as VirB/VirD4 and is composed of 12 subunits [25]. It consists of cytoplasmic ATPases (VirD4, VirB4, VirB11), which provide energy for the translocation process and are the first to bind to effector molecules [36], inner (VirB3, VirB6, VirB8) and outer (VirB7, VirB9, VirB10) membrane components forming the transmembrane channel [24], as well as structural pilus proteins (VirB1, VirB2, VirB5) [37]. This secretion machinery successfully transports tumor-inducing Ti-plasmid fragments into eukaryotic plant cells [25] and a number of auxiliary proteins [38, 39]. In plants, both groups of molecules promote the development of tumor-like growths (galls) where the further proliferation of bacterial cells takes place [25]. T4SS-A is associated not only with the virulence of phytopathogens [40], but also with the virulence of human pathogens. For example, in Helicobacter pylori, T4SS-A effector molecules induce a pro-inflammatory response in gastric epithelial cells [41] and cause uncontrolled division of host cells [42].
Unlike A. tumefaciens T4SS-A, class B secretion systems deliver primarily protein effectors to eukaryotic target cells [43] and play a critical role in the pathogenesis of such diseases as Legionnaires' disease [26] and Q fever [44, 45]. Among T4SSs-B in gram-negative bacteria, the most extensively studied secretion machinery is Dot/Icm in Legionella pneumophila – an intracellular pathogen and the causative agent of legionellosis. It participates in the transport of more than 300 effector molecules, most of which have not been described yet. Effector molecules are involved in formation of special replicative phagosomes in eukaryotic cells [46] and interfere with vital activities of the host cell [47, 48], promoting intracellular proliferation of legionella [49].
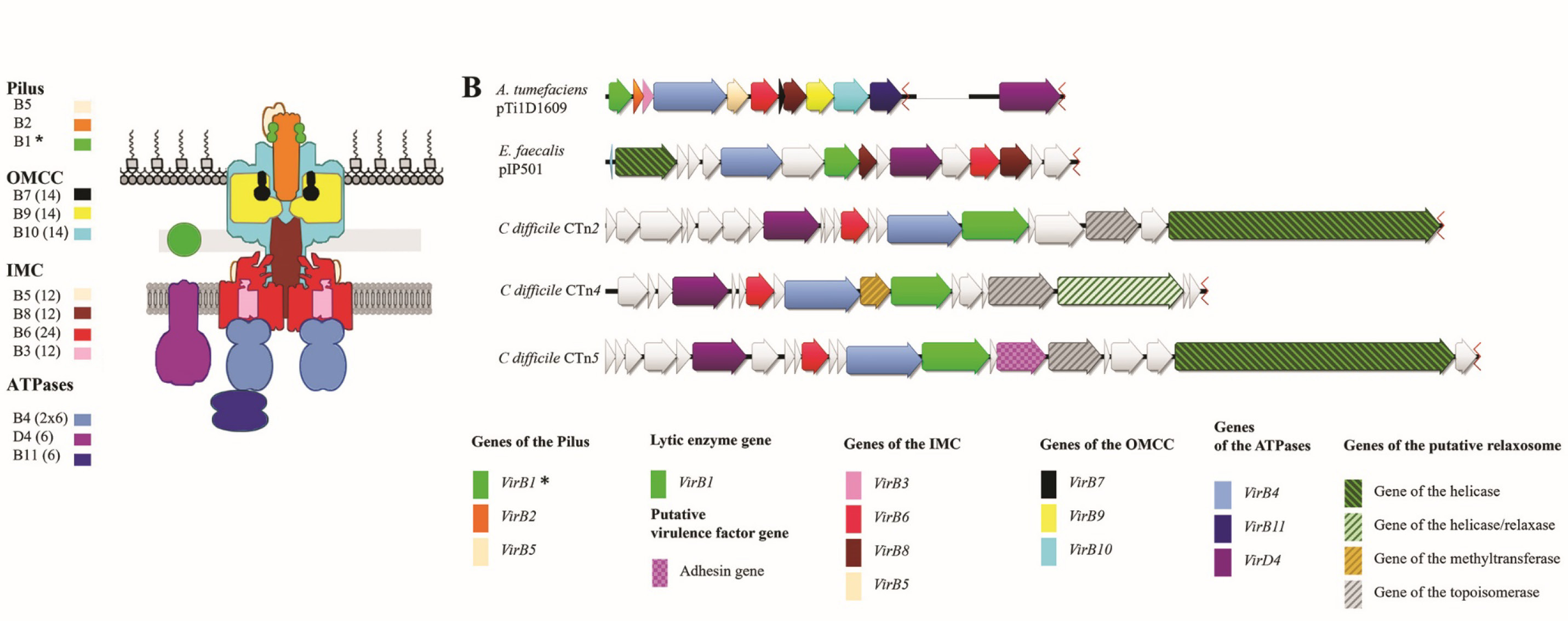
Conjugative T4SSs-C in gram-positive microorganisms are similar in structure to the above systems and include homologs of the respective subunits [33]. For example, the Enterococcus sp. plasmid pIP501 (Fig. 1, b) contains:
- TraA relaxase (required for formation of a single-stranded DNA fragment);
- TraG hydrolase (a VirB1 homolog), which cleaves bonds in peptidoglycan during the formation of the secretion machinery;
- proteins forming a channel through the cell wall – TraL (a VirB6 homolog), TraM, and TraH (the last two are putative VirB8 homologs);
- TraE and TraJ ATPases (VirB4 and VirD4 homologs, respectively) [50–52].
As the outer membrane is absent in gram-positive microorganisms, the T4SS-C secretion machinery can be represented by the so-called "minimized" structures comprising only 4–7 subunits [34, 35] compared to 12 subunits in A. tumefaciens [25] or 27 in L. pneumophila [53].
While in gram-negative bacteria T4SS is strongly associated with the pathogenesis of infections, in gram-positive bacteria this type of secretion machinery is primarily seen as a participant in conjugative processes and, consequently, as a factor in the dissemination of antibiotic resistance genes (Clostridium perfringens plasmids pCW3, Enterococcus sp. plasmids pIP501, and others) [51, 52, 54]. In the meantime, it can be assumed that, like A. tumefaciens and H. pylori secretion systems, gram-positive bacteria can use the T4SS machinery both for delivery of single-stranded DNA molecules and for translocation of molecules promoting development of infectious processes; this assumption has already been supported by findings of some researchers. It has been found that adhesins secreted by T4SS-C of E. faecalis plasmid pCF10 promote the formation of biofilms and increase the enterococcal virulence [30]. Another example of T4SS-C participation in the pathogenesis of infectious diseases can be found in the data on the association between the Sp1 genomic pathogenicity island of Streptococcus suis and the outbreaks of toxic shock syndrome in 1998 and 2005 [55, 56]. Sp1, which is responsible for adaptive and virulent properties of the streptococcus, contains homologs of only 4 T4SS genes: VirB1, VirB4, VirD4, and VirB6 [56, 57]. Despite its simple structure, this minimized secretion system retains its functional characteristics and is not only involved in the conjugative transfer of the pathogenicity island, but also plays a direct role in the virulence of S. suis [28, 29]. Thus, the secretion of protein virulence factors and translocation of T4SS-C MGEs provide evidence of the participation of this machinery in the pathogenesis of infections caused by gram-positive microorganisms, as it has been shown earlier for gram-negative bacteria.
Class C type 4 secretion system in C. difficile
Despite its potential significance in the pathogenesis of infection, T4SS-C in C. difficile is still poorly studied [58, 24]. The detailed structure of the secretion machinery has not been identified so far. In addition to VirD4, VirB6, and VirB4 genes [24], which were identified using bioinformatics analysis, we discovered a gene presumably encoding a VirB1 homolog in all three conjugative transposons: CTn4, CTn2, and CTn5 (the last two are referred to as CTn2/CTn5 in this article) [31]. In C. difficile, the conjugative part of CTn operons also includes methyltransferase, relaxase, helicase, and topoisomerase genes. Most likely, they encode components of relaxosome (Fig. 1, b), which is required for formation of transportable single-stranded DNA molecules and for DNA transfer to the secretion machinery similar to the process involving the A. tumefaciens VirD2 protein [25] and the TraA relaxase of plasmid pIP501 [51]. It is still unclear whether the gene product from the conjugative transposon, CD630_18580, belongs to T4SS-C, as, according to Bhatty et al. [34], it is an ortholog of E. faecalis adhesin pCF10 [30] and, by analogy with the enterococcal protein, can act as a virulence factor in C. difficile.
Fig. 1. T4SS organization.
a — schematic representation of the Agrobacterium tumefaciens type 4A secretion machinery [25]; b — T4SS-C components of three conjugative transposons of C. difficile strain 630 compared to representatives of T4SS-A (A. tumefaciens pTi1D1609) and T4SS-C (Enterococcus faecalis pIP501).
The visualization was performed using the Genious software; the data on each component were verified using NCBI and UniProt databases, as well as using separate pairwise alignments. IMC — the inner membrane complex; OMCC — the outer membrane core complex. *In T4SS-A, VirB1, being a lytic enzyme, breaks bonds within the peptidoglycan during the formation of a transmembrane channel; it also forms a part of the pilus assembly.
VirB4 and VirD4 proteins are the most important components of bacterial T4SS; they take part in translocation as an energy source for transport of biomolecules. Both ATPases belong to conserved proteins of the secretion machinery [58, 59] and, consequently, are the best targets for taxonomic studies [60]. The phylogenetic analysis of sequences of VirB4 and VirD4-like ATPases makes it possible to identify which class (A, B, or C) T4SS including the above enzymes should be assigned to (Fig. 2). Because of the differences in their structure, C. difficile VirB4 and VirD4 of transposons CTn4 and CTn2/CTn5 fall into different clades, which, together with ATPases of the pathogenicity island of S. suis constitute three taxonomically important groups within T4SS-C, thus implying that their functional significance can be different.
Fig. 2. Phylogenetic trees of VirD4- (a) and VirB4-like (b) proteins of gram-positive and gram-negative bacteria.
The green arrow indicates CTn4 ATPases; the orange arrow indicates CTn2/5. The alignment was performed using MAFFT and K-align tools; trees were constructed using the maximum likelihood method and the Blosum62 matrix [31].
The only biochemical research addressing the clostridial T4SS mechanism and the above proteins has been recently conducted in our laboratory [31]. Based on our findings, both enzymes have an Mg2+-dependent ATPase activity, and the maximum rate of the catalytic reaction is reached in the presence of potassium ions [31]. VirD4, but not VirB4 can interact with nucleic acid molecules. An important role in this interaction belongs to tryptophan at amino acid position 241, since the W241A point substitution results in a protein variant that is unable to adsorb DNA [31]. VirB4 and VirD4 form enzymatically active oligomeric complexes, while the substitution of the key amino acids in the so-called Walker A and Walker B motifs within enzyme domains not only decreases the ATPase activity, but also destabilizes the entire oligomeric complex [31]. The similarity between both ATPases and other T4SS ATPases in the amino acid composition (Fig. 2), biochemical and structural characteristics [31] suggests that this secretion machinery can translocate proteins like the secretion system of S. suis and E. faecalis. However, this hypothesis requires further research.
The presence of VirB4 and VirD4-like ATPases in strain 630 is neither unique nor rare. The frequency of occurrence of T4SS-C ATPases among representatives of C. difficile can be efficiently assessed using sequenced annotated genomes assembled into chromosomes. For the C. difficile species, a total of 17,961 sequenced genomes were deposited to the NCBI database by the end of 20221; 92 of them were annotated and assembled to the chromosome level, while 89 genomes, when duplicate variants are discarded, fall within the set parameters (Fig. 3). Half of such genomes (45 strains) contain T4SS-C ATPases in conjugative transposons; these amino acid sequences were not found in the other genomes using the Blast algorithm [Sorokina et al., unpublished data]. In 38 strains, the secretion machinery genes are not damaged (Fig. 3), thus being most likely functionally active. Only C. difficile 630 has three variants of genes of VirB4 and VirD4 ATPases, which are located in transposons CTn2, CTn4 and CTn5. In 10 genomes, both VirB4 and VirD4 were found in 2 loci (transposons), and, most frequently, each gene is represented by one copy. T4SS ATPases demonstrating low homology with ATPases of the known transposons CTn4 or CTn2/CTn5 (less than 80% of identity) are of special interest and require further research. By their structure, T4SSs-C that include the similar "new" variants of ATPases are highly similar to the systems in CTn4 or CTn2/CTn5, except for individual cases having the gene with the motifs typical of cell wall proteins (potential virulence factors) between topoisomerase and helicase genes [61, 62]. In the subset of this size, it is impossible to estimate the homogeneity of the latter group. On the whole, our taxonomic analysis makes it possible not only to identify strains with ATPases, which belong to the known subgroups (CTn4 or CTn2/CTn5), but also to detect new variants of the secretion machinery.
Fig. 3. Distribution of VirB4 and VirD4 T4SS-C ATPases among C. difficile strains with annotated genomes in the NCBI database.
In genomes, ATPases could be present in 1 (1 locus), 2 (2 loci), or 3 (3 loci) variants. If amino acid sequences were more than 87% identical to WP_011861117.1 (VirD4Ctn4) and WP_011861114.1 (VirB4Ctn4), we assigned them to the CTn4 group. If both sequences were more than 87% identical to WP_011860784.1 (VirD4Ctn2/5) and WP_042741540.1 (VirB4Ctn2/5), they were assigned to the CTn2/5 group. At lower values, ATPases were assigned to the CTnX group. The amino acid sequences lacking Walker A or Walker B motifs, which participate in the formation of oligomeric complexes and constitute the active center of the enzyme, were seen as damaged.
Conclusion
C. difficile infection remains a serious problem; the increasing frequency of nosocomial outbreaks and the emergence of community-acquired forms heighten the need for new prevention and treatment methods. Based on the findings of recent studies, we can conclude that MGEs contribute to high virulence difficile. T4SSs are significant components of MGEs; their impressive diversity in gram-positive microorganisms in general and in C. difficile, in particular, implies their high evolutionary and, consequently, medical significance. Initially capable of transferring genes involved in the adaptive response to adverse environmental factors, later on, T4SSs acquired the ability to transport protein molecules – virulence factors. These processes are convincingly described in publications addressing secretion systems in pathogenic gram-negative microorganisms. In gram-positive bacteria, specifically in C. difficile, this type of MGE participation in the pathogenesis of infectious diseases has been much more poorly studied. The composition of the secretion machinery has not been identified. In addition to the above VirB4 and VirD4 ATPases, as well as the VirB6 transmembrane channel protein, the secretion machinery may include the VirB1 homolog, relaxosome components, and adhesins. The latter, representing potential virulence factors, are of particular interest. Further studies of the composition and structure of entire T4SS-C as well as its individual components can significantly contribute to the progress in understanding the pathogenesis of the respective infections and help develop pathogenically grounded approaches to prevention and treatment of diseases caused by C. difficile.
Funding source. This study was not supported by any external sources of funding.
Conflict of interest. The authors declare no apparent or potential conflicts of interest related to the publication of this article.
Author contribution. Аll authors made a substantial contribution to the conception of the work, acquisition, analysis, interpretation of data for the work, drafting and revising the work, final approval of the version to be published.
1 URL: https://www.ncbi.nlm.nih.gov/data-hub/taxonomy/1496/
About the authors
Julya V. Sorokina
Gamaleya Research Centre for Epidemiology and Microbiology
Author for correspondence.
Email: YV_Sorokina@gamaleya.org
ORCID iD: 0000-0002-1869-742X
Researcher, Department of Bacterial Infections
Russian Federation, MoscowYuriy F. Belyi
Gamaleya Research Centre for Epidemiology and Microbiology
Email: belyi@gamaleya.org
ORCID iD: 0000-0003-2312-1465
D. Sci. (Med.), Head, Department of Bacterial Infections
Russian Federation, MoscowReferences
- Hall I.C., O'Toole E. Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. Am. J. Dis. Child. 1935;49(2):390. DOI: https://doi.org/10.1001/archpedi.1935.01970020105010
- Riley T.V., Wymer V., Bamford V.W., Bowman R.A. Clostridium difficile in general practice and community health. J. Hyg. (Lond.). 1986;96(1):13–7. DOI: https://doi.org/10.1017/s0022172400062483
- Guh A.Y., Mu Y., Winston L.G., et al. Trends in U.S. Burden of Clostridioides difficile infection and outcomes. N. Engl. J. Med. 2020;382(14):1320–30. DOI: https://doi.org/10.1056/nejmoa1910215
- Lyerly D.M., Saum K.E., MacDonald D.K., Wilkins T.D. Effects of Clostridium difficile toxins given intragastrically to animals. Infect. Immun. 1985;47(2):349–52. DOI: https://doi.org/10.1128/iai.47.2.349-352.1985
- Orrell K.E., Melnyk R.A. Large clostridial toxins: mechanisms and roles in disease. Microbiol. Mol. Biol. Rev. 2021;85(3):e0006421. DOI: https://doi.org/10.1128/mmbr.00064-21
- Popoff M.R., Rubin E.J., Gill D.M., Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect. Immun. 1988;56(9):2299–306. DOI: https://doi.org/10.1128/iai.56.9.2299-2306.1988
- Aktories K., Papatheodorou P., Schwan C. Binary Clostridium difficile toxin (CDT) — a virulence factor disturbing the cytoskeleton. Anaerobe. 2018;53:21–9. DOI: https://doi.org/10.1016/j.anaerobe.2018.03.001
- Riedel T., Neumann-Schaal M., Wittmann J., et al. Characterization of Clostridioides difficile DSM 101085 with A–B–CDT + phenotype from a late recurrent colonization. Genome Biol. Evol. 2020;12(5):566–77. DOI: https://doi.org/10.1093/gbe/evaa072
- Merrigan M.M., Venugopal A., Roxas J.L., et al. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS One. 2013;8(11):e78404. DOI: https://doi.org/10.1371/journal.pone.0078404
- Shen A. Clostridioides difficile spore formation and germination: new insights and opportunities for intervention. Annu. Rev. Microbiol. 2020;74(1):545–66. DOI: https://doi.org/10.1146/annurev-micro-011320-011321
- Taggart M.G., Snelling W.J., Naughton P.J., et al. Biofilm regulation in Clostridioides difficile: novel systems linked to hypervirulence. PLOS Pathog. 2021;17(9):e1009817. DOI: https://doi.org/10.1371/journal.ppat.1009817
- de la Riva L., Willing S.E., Tate E.W., Fairweather N.F. Roles of cysteine proteases Cwp84 and Cwp13 in biogenesis of the cell wall of Clostridium difficile. J. Bacteriol. 2011;193(13):3276–85. DOI: https://doi.org/10.1128/jb.00248-11
- Coullon H., Candela T. Clostridioides difficile peptidoglycan modifications. Curr. Opin. Microbiol. 2022;65:156–61. DOI: https://doi.org/10.1016/j.mib.2021.11.010
- McKee R.W., Harvest C.K., Tamayo R. Cyclic diguanylate regulates virulence factor genes via multiple riboswitches in Clostridium difficile. mSphere. 2018;3(5):e00423-18. DOI: https://doi.org/10.1128/msphere.00423-18
- Buddle J.E., Fagan R.P. Pathogenicity and virulence of Clostridioides difficile. Virulence. 2023;14(1):2150452. DOI: https://doi.org/10.1080/21505594.2022.2150452
- Androga G.O., Knight D.R., Hutton M.L., et al. In silico, in vitro and in vivo analysis of putative virulence factors identified in large clostridial toxin-negative, binary toxin-producing C. difficile strains. Anaerobe. 2019;60:102083. DOI: https://doi.org/10.1016/j.anaerobe.2019.102083
- Lee A.S.Y., Song K.P. LuxS/autoinducer-2 quorum sensing molecule regulates transcriptional virulence gene expression in Clostridium difficile. Biochem. Biophys. Res. Commun. 2005;335(3):659–66. DOI: https://doi.org/10.1016/j.bbrc.2005.07.131
- Okada Y., Okugawa S., Ikeda M., et al. Genetic diversity and epidemiology of accessory gene regulator loci in Clostridioides difficile. Access Microbiol. 2020;2(7):acmi.0.000134. DOI: https://doi.org/10.1099/acmi.0.000134
- Ahmed U.K.B., Shadid T.M., Larabee J.L., Ballard J.D. Combined and distinct roles of Agr proteins in Clostridioides difficile 630 sporulation, motility, and toxin production. mBio. 2020;11(6):e03190-20. DOI: https://doi.org/10.1128/mbio.03190-20
- Aguilar-Zamora E., Weimer B.C., et al. Molecular epidemiology and antimicrobial resistance of Clostridioides difficile in hospitalized patients from Mexico. Front. Microbiol. 2022;12:787451. DOI: https://doi.org/10.3389/fmicb.2021.787451
- Wen X., Shen C., Xia J., et al. Whole-genome sequencing reveals the high nosocomial transmission and antimicrobial resistance of Clostridioides difficile in a single center in China, a four-year retrospective study. 2021;10(1):e01322-21. DOI: https://doi.org/10.1128/spectrum.01322-21
- Darkoh C., Keita K., Odo C., et al. Emergence of clinical Clostridioides difficile isolates with decreased susceptibility to vancomycin. Clin. Infect. Dis. 2022;74(1):120–6. DOI: https://doi.org/10.1093/cid/ciaa912
- Launay A., Ballard S.A., Johnson P.D., et al. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob. Agents Chemother. 2006;50(3):1054–62. DOI: https://doi.org/10.1128/aac.50.3.1054-1062.2006
- Zhang W., Cheng Y., Du P., et al. Genomic study of the type IVC secretion system in Clostridium difficile: understanding C. difficile evolution via horizontal gene transfer. Genome. 2017;60(1):8–16. DOI: https://doi.org/10.1139/gen-2016-0053
- Li Y.G., Christie P.J. The agrobacterium VirB/VirD4 T4SS: mechanism and architecture defined through in vivo mutagenesis and chimeric systems. Curr. Top. Microbiol. Immunol. 2018;418:233–60. DOI: https://doi.org/10.1007/82_2018_94
- Böck D., Hüsler D., Steiner B., et al. The polar Legionella Icm/Dot T4SS establishes distinct contact sites with the pathogen vacuole membrane. mBio. 2021;12(5):e0218021. DOI: https://doi.org/10.1128/mbio.02180-21
- Grohmann E., Christie P.J., Waksman G., Backert S. Type IV secretion in gram-negative and gram-positive bacteria: type IV secretion. Mol. Microbiol. 2018;107(4):455–71. DOI: https://doi.org/10.1111/mmi.13896
- Zhao Y., Liu G., Li S., et al. Role of a type IV-like secretion system of Streptococcus suis 2 in the development of streptococcal toxic shock syndrome. J. Infect. Dis. 2011;204(2):274–81. DOI: https://doi.org/10.1093/infdis/jir261
- Zhong Q., Zhao Y., Chen T., et al. A functional peptidoglycan hydrolase characterized from T4SS in 89K pathogenicity island of epidemic Streptococcus suis serotype 2. BMC Microbiol. 2014;14:73. DOI: https://doi.org/10.1186/1471-2180-14-73
- Bhatty M., Cruz M.R., Frank K.L., et al. Enterococcus faecalis pCF10-encoded surface proteins PrgA, PrgB (aggregation substance), and PrgC contribute to plasmid transfer, biofilm formation, and virulence. Mol. Microbiol. 2015;95(4):660–77. DOI: https://doi.org/10.1111/mmi.12893
- Sorokina J., Sokolova I., Rybolovlev I., et al. VirB4- and VirD4-like ATPases, components of a putative type 4C secretion system in Clostridioides difficile. J. Bacteriol. 2021;203(21):e00359-21. DOI: https://doi.org/10.1128/jb.00359-21
- Backert S. Erratum to: Type IV secretion in Gram-negative and Gram-positive bacteria. Curr. Top. Microbiol. Immunol. 2017; 413:E1. DOI: https://doi.org/10.1007/978-3-319-75241-9_14
- Zhang W., Rong C., Chen C., Gao G.F. Type-IVC secretion system: a novel subclass of type IV secretion system (T4SS) common existing in gram-positive genus Streptococcus. PLoS One. 2012;7(10):e46390. DOI: https://doi.org/10.1371/journal.pone.0046390
- Bhatty M., Laverde Gomez J.A., Christie P.J. The expanding bacterial type IV secretion lexicon. Res. Microbiol. 2013;164(6):620–39. DOI: https://doi.org/10.1016/j.resmic.2013.03.012
- Wang J., Feng Y., Wang C., et al. Pathogenic Streptococcus strains employ novel escape strategy to inhibit bacteriostatic effect mediated by mammalian peptidoglycan recognition protein. Cell. Microbiol. 2017;19(7). DOI: https://doi.org/10.1111/cmi.12724
- Das A. Identification of a carboxy-terminal glutamine-rich domain in Agrobacterium tumefaciens coupling protein VirD4 required for recognition of T-Strand DNA and Not VirE2 as a substrate for transfer to plant cells. Mol. Plant Microbe Interact. 2020;33(2):166–72. DOI: https://doi.org/10.1094/mpmi-04-19-0099-r
- Amro J., Black C., Jemouai Z., et al. Cryo-EM structure of the Agrobacterium tumefaciens T-pilus reveals the importance of positive charges in the lumen. Structure. 2023;31(4):375–84.e4. DOI: https://doi.org/10.1016/j.str.2022.11.007
- Roushan M.R., de Zeeuw M.A.M., Hooykaas P.J.J., van Heusden G.P.H. Application of phiLOV2.1 as a fluorescent marker for visualization of Agrobacterium effector protein translocation. Plant J. 2018;96(3):685–99. DOI: https://doi.org/10.1111/tpj.14060
- Чумаков М.И., Мазилов С.И., Гусев Ю.С., Волохина И.В. Исследование способности агробактериального белка VirE2 к образованию пор в мембранах. Биологические мембраны. 2010;27(5):449–54. Chumakov M.I., Mazilov S.I., Gusev Yu.S., Volokhina I.V. Study of the ability of agrobacterial protein VirE2 to form pores in membranes. Biological Membranes. 2010;27(5):449–54. EDN: https://www.elibrary.ru/mvskun
- Дюбо Ю.В., Николайчик Е.А. Модификация вирулентных свойств Pectobacterium atrosepticum конъюгативной плазмидой pPA21A. Молекулярная и прикладная генетика. 2018;24:37–44. Dyubo Yu.V., Nikolaichik E.A. Modification virulent properties of Pectobacterium atrosepticum by conjugative plasmid PPA21A. Molecular and Applied Genetics. 2018;24:37–44. EDN: https://www.elibrary.ru/hgoxko
- Pfannkuch L., Hurwitz R., Traulsen J., et al. ADP heptose, a novel pathogen‐associated molecular pattern identified in Helicobacter pylori. FASEB J. 2019;33(8):9087–99. DOI: https://doi.org/10.1096/fj.201802555r
- Zhang X., Li C., Chen D., et al. H. pylori CagA activates the NLRP3 inflammasome to promote gastric cancer cell migration and invasion. Inflamm. Res. 2022;71(1):141–55. DOI: https://doi.org/10.1007/s00011-021-01522-6
- Allombert J., Jaboulay C., Michard C., et al. Deciphering Legionella effector delivery by Icm/Dot secretion system reveals a new role for c-di-GMP signaling. J. Mol. Biol. 2021;433(13):166985. DOI: https://doi.org/10.1016/j.jmb.2021.166985
- Beare P.A., Gilk S.D., Larson C.L., et al. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio. 2011;2(4):e00175-11. DOI: https://doi.org/10.1128/mbio.00175-11
- Clemente T.M., Augusto L., Angara R.K., Gilk S.D. Coxiella burnetii actively blocks IL-17-induced oxidative stress in macrophages. bioRxiv. 2023;2023.03.15.532774. Preprint. DOI: https://doi.org/10.1101/2023.03.15.532774
- Luo J., Wang L., Song L., Luo Z.Q. Exploitation of the host ubiquitin system: means by Legionella pneumophila. Front. Microbiol. 2021;12:790442. DOI: https://doi.org/10.3389/fmicb.2021.790442
- Тартаковская Д.И. Эукариотические мишени цитотоксической глюкозилтрансферазы Legionella pneumophila. Инфекция и иммунитет. 2012;2(1-2):325. Tartakovskaya D.I. Eukaryotic targets of cytotoxic glucosyltransferase Legionella pneumophila. Russian Journal of Infection and Immunity. 2012;2(1-2):325.
- Levanova N., Steinemann M., Böhmer K.E., Schneider S., et al. Characterization of the glucosyltransferase activity of Legionella pneumophila effector SetA. Naunyn Schmiedebergs Arch. Pharmacol. 2019;392(1):69–79. DOI: https://doi.org/10.1007/s00210-018-1562-9
- Liu L., Roy C.R. The Legionella pneumophila effector RavY contributes to a replication-permissive vacuolar environment during infection. Infect. Immun. 2021;89(12):e0026121. DOI: https://doi.org/10.1128/iai.00261-21
- Kurenbach B., Bohn C., Prabhu J., et al. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid. 2003;50(1):86–93. DOI: https://doi.org/10.1016/s0147-619x(03)00044-1
- Abajy M.Y., Kopeć J., Schiwon K., et al. A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in Gram-positive bacteria. J. Bacteriol. 2007;189(6):2487–96. DOI: https://doi.org/10.1128/jb.01491-06
- Kohler V., Vaishampayan A., Grohmann E. Broad-host-range Inc18 plasmids: occurrence, spread and transfer mechanisms. Plasmid. 2018;99:11–21. DOI: https://doi.org/10.1016/j.plasmid.2018.06.001
- Durie C.L., Sheedlo M.J., Chung J.M., et al. Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex. eLife. 2020;9:e59530. DOI: https://doi.org/10.7554/elife.59530
- Revitt-Mills S.A., Watts T.D., Lyras D., et al. The ever-expanding tcp conjugation locus of pCW3 from Clostridium perfringens. Plasmid. 2021;113:102516. DOI: https://doi.org/10.1016/j.plasmid.2020.102516
- Tang J., Wang C., Feng Y., et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 2006; 3(5):e151. DOI: https://doi.org/10.1371/journal.pmed.0030151
- Li M., Wang C., Feng Y., et al. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS One. 2008;3(5):e2080. DOI: https://doi.org/10.1371/journal.pone.0002080
- Li M., Shen X., Yan J., et al. GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2: T4SS-mediated transfer of 89K PAI in S. suis 2. Mol. Microbiol. 2011;79(6):1670–83. DOI: https://doi.org/10.1111/j.1365-2958.2011.07553.x
- Li N., Jia H., Yang H., et al. Preliminary screening of type IV secretion system in divergent geographic sources of Clostridium difficile. Exp. Ther. Med. 2017;14(5):4405–10. DOI: https://doi.org/10.3892/etm.2017.5065
- Whitaker N., Berry T.M., Rosenthal N., et al. Chimeric coupling proteins mediate transfer of heterologous type IV effectors through the Escherichia coli pKM101-encoded conjugation machine. J. Bacteriol. 2016;198(19):2701–18. DOI: https://doi.org/10.1128/jb.00378-16
- Fernández-López R., Garcillán-Barcia M.P., Revilla C., et al. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 2006;30(6):942–66. DOI: https://doi.org/10.1111/j.1574-6976.2006.00042.x
- Tulli L., Marchi S., Petracca R., et al. CbpA: a novel surface exposed adhesin of Clostridium difficile targeting human collagen: сollagen binding protein of Clostridium difficile. Cell. Microbiol. 2013;15(10):1674–87. DOI: https://doi.org/10.1111/cmi.12139
- Malik A., Shoombuatong W., Kim C.B., Manavalan B. GPApred: The first computational predictor for identifying proteins with LPXTG-like motif using sequence-based optimal features. Int. J. Biol. Macromol. 2023;229:529–38. DOI: https://doi.org/10.1016/j.ijbiomac.2022.12.315
Supplementary files