Virulence and tissue tropism of different epidemiologically significant SARS-CoV-2 variants for golden Syrian hamsters
- Authors: Gracheva A.V.1, Drokov A.O.1, Smirnova D.I.1, Khokhlova D.M.1, Korchevaya E.R.1, Pankratov A.A.2, Trunova G.V.2, Khokhlova V.A.2, Vorontsova M.S.2, Leneva I.A.1, Svitich O.A.1,3, Zverev V.V.1,3, Faizuloev E.B.1,4
-
Affiliations:
- I.I. Mechnikov Research Institute for Vaccines and Sera
- P.A. Hertsen Moscow Oncology Research Institute — the branch of the National Medical Research Radiological Center
- I.M. Sechenov First Moscow State Medical University (Sechenov University)
- Russian Medical Academy of Continuous Professional Education
- Issue: Vol 101, No 4 (2024)
- Pages: 470-482
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18645
- DOI: https://doi.org/10.36233/0372-9311-528
- EDN: https://elibrary.ru/jukmca
- ID: 18645
Cite item
Abstract
Introduction. Animal models for SARS-CoV-2 infection, reproducing the clinical features of COVID-19 in humans, are important tools for studying the pathogenesis of the disease, transmission of the pathogen and are indispensable for testing antiviral drugs and vaccines.
The aim of the study was to assess the virulence and tissue tropism for golden Syrian hamsters of SARS-CoV-2 strains belonging to different epidemiologically significant variants: Wuhan-like, Delta, Omicron BA.1.1 and Omicron BA.5.2.
Materials and methods. Hamsters were intranasally infected with different SARS-CoV-2 strains. Virulence and tissue tropism of SARS-CoV-2 strains were assessed by comparing the dynamics of weight, viral load in organs and histopathological changes in lungs in infected and uninfected animals.
Results. The Wuhan-like Dubrovka strain had the greatest virulence for hamsters, which was manifested by the development of severe pneumonia and a delay in weight gain by 14.6%, high virus content in the lungs, nasal passages and brain — 6.2, 5,9 and 3.7 lg TCID50/ml of homogenate, respectively. Presumably, it was the infection of the Wuhan-like virus of the central nervous system that negatively affected the weight and general condition of the animals. When hamsters were infected with viruses belonging to the Delta and Omicron variants, the observed minor weight loss in animals was uninformative, so indicators such as lung histopathology, viral load in the lungs, nasal passages, heart and other organs played a decisive role in assessing the virus pathogenicity. A score assessment of lung histopathology was of particular value in assessing the severity of pneumonia, since it reduced subjectivity in evaluating the results of histological examination and provided a semi-quantitative assessment of the pathological process.
Conclusion. Despite the revealed lower virulence for hamsters of viruses belonging to the Delta and Omicron variants compared to the ancestral Wuhan virus, this animal model for COVID-19 retains its value for conducting preclinical trials of antiviral drugs.
Full Text
Introduction
Modeling viral diseases in laboratory animals is one of the most important problems of medical virology. The emergence in 2019 and global spread of SARS-CoV-2 coronavirus (Severe acute respiratory syndrome-related coronavirus species, Betacoronavirus genus, Coronaviridae family), accompanied by a high rate of hospitalization and mortality among those who became ill, necessitated the urgent development of treatments and specific prophylaxis for COVID-19, which is impossible without preclinical testing in adequate animal models of the disease. Since the beginning of the pandemic, considerable efforts have been made to develop effective and safe vaccines and therapeutic agents, and studies of the pathogenesis and features of the immune response to SARS-CoV-2 infection have been conducted [1]. The success of these studies largely depended on the availability of animal models for coronavirus infection developed in the first decade of the 2000s against the background of the threat of worldwide spread of SARS-CoV-1, the causative agent of severe acute respiratory syndrome [2], which belongs to the same species as SARS-CoV-2. Animal models for infection that reproduce clinical and pathological features of COVID-19 in humans are important tools for studying the pathogenesis of the disease, pathogen transmission and are indispensable for testing new antiviral drugs and vaccines [3–5].
To date, there are several animal models for COVID-19, primarily based on representatives of primate, carnivore and rodent groups. However, the problem of selecting the most adequate, informative and convenient model remains relevant. The value of primate-based animal models for coronavirus pneumonia lies in the fact that monkeys are similar to humans in their physiological characteristics and immune regulation. Rhesus macaques, African green monkeys, baboons, and common marmosets are most often used for COVID-19 modeling [1, 6, 7]. The main drawbacks of such models are the huge demand for animals, high cost, shortage of trained personnel and primate vivariums equipped according to biosafety level 3 requirements [1, 6].
Mink, ferrets and cats are also susceptible to SARS-CoV-like coronaviruses [7-11]. Notably, SARS-CoV-2 is found in the nasal cavity of ferrets and they can be infected by indirect contact, indicating the ability of ferrets and mink to transmit the virus by mimicking the SARS-CoV-2 transmission pathway in humans. A disadvantage of such models is that these animals are relatively large carnivores, so handling them is difficult. Therefore, there is a need for models based on small laboratory animals that are susceptible to the virus.
Mice and other rodents are most commonly used to model COVID-19. However, wild-type mice are not susceptible to infection with the ancestral Wuhan-like virus SARS-CoV-2 [5, 6, 12] because the virus is able to bind efficiently to the human ACE2 receptor (hACE2) but not to murine ACE2 (mACE2). Our previous findings indicate that Wuhan-like virus does not cause productive infection in BALB/c mice and, in contrast, when infected with Omicron-like virus multiplies in lungs, brain tissue, and other organs [5].
Several lines of genetically modified mice with hACE2 receptor are known, which have been adapted for studies on the pathogenesis of cardiovascular diseases and modeling of coronavirus infection [6, 12]. These transgenic mouse lines, with different origin, are capable of stable hACE2 expression in many organs. The mouse model also has some serious limitations, including differences in hACE2 expression patterns in different organs and tissues in humans and mice. Because hACE2 expression in transgenic mice is not physiologic, infection with SARS-CoV-2 can cause clinical manifestations and pathologic changes uncharacteristic of humans [1]. In addition, transgenic mice are not widely available in Russia and are characterized by high cost.
Among SARS-CoV-2 susceptible animals, the golden Syrian hamster (Mesocricetus auratus; hereafter hamsters) is of particular interest. Genetic comparison of hACE2 with analogous receptors of other mammals showed that the amino acid sequence of hamster ACE2 is very similar to that of the analogous human receptor, with which it has only 3-4 differences. In addition, hamster ACE2 has shown high affinity for the S-protein of SARS-CoV-2 and SARS-CoV in several studies [1, 3, 7].
The hamster model for coronavirus pneumonia is widely used in preclinical studies of vaccines and drugs [13]. Symptoms, disease pathogenesis and immune responses characteristic of COVID-19 in humans are well reproduced in hamsters [3, 14]. Hamsters are also in demand for modeling other human respiratory viral infections [14] caused by viruses such as SARS-CoV-1 [2], influenza viruses [15, 16] and adenoviruses [14, 17]. With advantages such as high reproduction rate, easy handling, affordable cost and availability in nurseries, hamsters are an optimal choice compared to other small laboratory animals.
Coronavirus disease caused in hamsters by Wuhan-like strains of SARS-CoV-2 has been well studied and characterized to date [13, 18-20]. Since at the current stage of the epidemic process, the ancestral SARS-CoV-2 virus has been replaced by new variants of concern (first Delta, then Omicron and its progeny), it is of interest to study their virulence and disease pathogenesis in infected hamsters. Previously, we conducted a study of the protective activity of a prototype live attenuated vaccine against SARS-CoV-2 in hamsters, which included their infection not only with the parental Wuhan-type virus, but also with strains belonging to Delta and Omicron variants [21]. In this article, we considered it appropriate to review and discuss the results obtained in more detail in the context of the pathogenicity of different virus variants for non-immunized hamsters.
The aim of the study was to assess the virulence and tissue specificity of SARS-CoV-2 strains belonging to different variants of concern in Syrian golden hamsters.
Materials and methods
Virus
Laboratory strains of SARS-CoV-2 isolated at the I.I. Mechnikov Research Institute of Virus Diseases from patients with confirmed diagnosis of COVID-19 during different periods of the pandemic were used in the study (Table 1). All works with SARS-CoV-2 virus were conducted in the conditions of the biosafety level 3 laboratory.
Table 1. Characteristics of SARS-CoV-2 strains used in the study
Strain | Collection date | GenBank ID | Variant | Pangolin lineage | Passage level | Titer, log10 TCID50/ml |
Dubrovka | 04.06.2020 | MW514307.1 | Wuhan-like | B.1.1.317 | 17 | 7,85 |
Podolsk | 10.08.2021 | ON032860.1 | Delta | AY.122 | 16 | 7,0 |
Otradnoe | 25.01.2022 | ON032857.1 | Omicron | BA.1.1 | 8 | 6,0 |
FEB2 | 11.10.2022 | OP920753.1 | Omicron | BA.5.2 | 4 | 6,5 |
SARS-CoV-2 was cultured in Vero CCL81 (ATCC) kidney epithelial cell culture of African green monkey (hereinafter referred to as Vero) at 37⁰С in DMEM medium based on Earle's buffer (PanEco) with 5% fetal bovine serum (Gibco), 300 μg/mL L-glutamine (PanEco), 40 μg/mL gentamicin (PanEco) in an atmosphere of 5% CO2. A three-day-old monolayer of Vero cells was infected with SARS-CoV-2 virus at a multiplicity of infection MOI = 0.001. Virus adsorption was performed in a CO2 incubator for 60 min, then maintenance medium (DMEM, 300 μg/mL L-glutamine, 40 μg/mL gentamicin) was added and incubated at 37⁰C until the appearance of pronounced cytopathic action (CPA) in an atmosphere of 5% CO2. After the appearance of pronounced CPA, the culture fluid was clarified by centrifugation at 4000 rpm for 10 min and stored at -80⁰C until used in experiments.
The titer of SARS-CoV-2 was determined in Vero cell culture by CPA endpoint. Tenfold dilutions of virus in 4 repeats were added to the wells of a 96-well plate with a 3-day-old monolayer of Vero cells and incubated for 5 days at 37⁰C in an atmosphere of 5% CO2. Titration results were evaluated by microscopic examination of the cell monolayer for the presence of characteristic CPA (rounding of cells and detachment of cells from the monolayer). Virus titer was calculated as described by M.A. Ramakrishnan et al. [22], and expressed in log10 TCID50/mL.
Animal models
4-week-old female SPF hamsters (n = 30) weighing 40-45 g (SPP Nursery for Laboratory Animals, Branch of the Institute of Bioorganic Chemistry of the RAS, Russia) were used in this work. Hamsters were randomly distributed into groups. The animals were kept in accordance with the rules for the arrangement, equipment and maintenance of experimental and biological clinics. The animals were fed with briquetted feed according to the approved norms. The authors complied with institutional and national standards for the use of laboratory animals in conducting the experimental animal study. The conduct of the study was approved by the Ethical Committee of the I.I. Mechnikov Research Institute of Veterinary Medicine (protocol No. 2 of 24.05.2021).
Study design
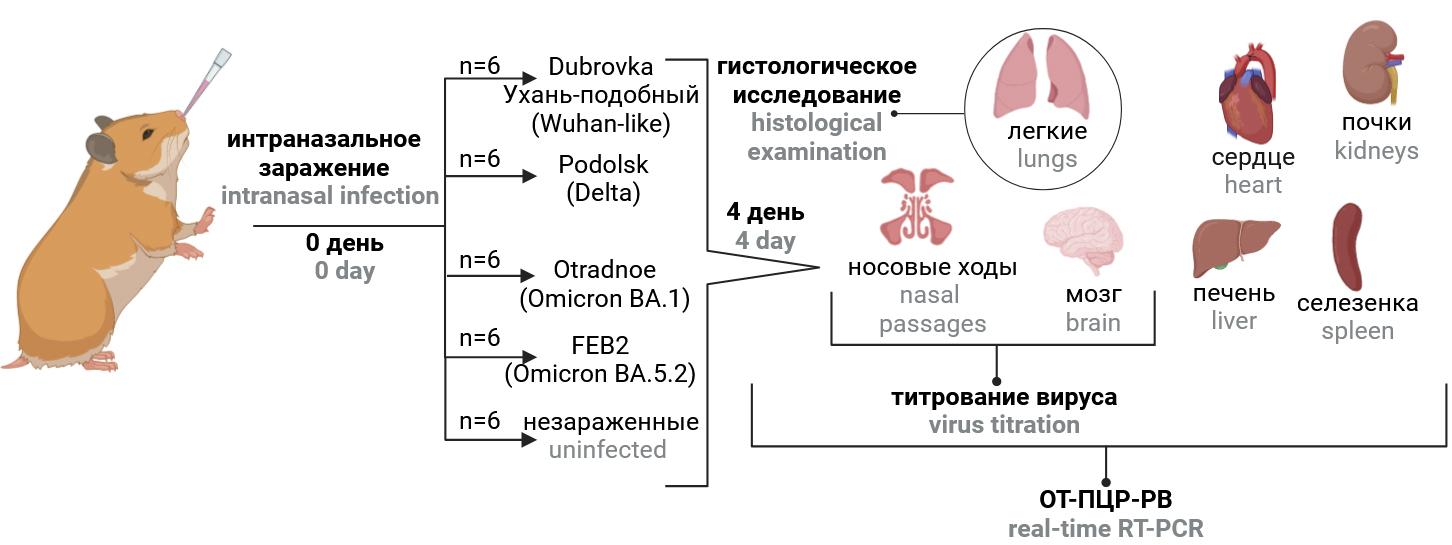
The study design is schematically presented in Fig. 1. Hamsters were divided into 5 groups of 6 animals each and intranasally infected with different virus strains (Table 1) at a dose of 104 TCID50/head (100 μl each). For intranasal infection, animals were anesthetized and held in an upright position. The negative control group received an equivalent volume of phosphate-salt buffer pH 7.2. Weight control was performed daily. Four days after infection, the animals were humanely euthanized. The right hamster lung was fixed in 10% neutral buffered formalin for histologic examination. Tissues of lung, brain, nasal passages, heart, liver, spleen, kidney and blood were collected, homogenized in 1 ml DMEM medium with gentamicin (40 μg/ml, PanEco) using a Tissue Lyser LT homogenizer (Qiagen) and centrifuged at 10,000 rpm for 5 min at 4ºC. The supernatant was collected for measurement of virus titers and viral RNA concentration and stored at –80ºC until examination. Changes in body weight from day 1 to day 4 after infection, virus titer and viral RNA content in organs and tissues, and severity of inflammatory changes in the lungs of animals on day 4 after infection reflected the virulence of the strain, and the distribution of viral RNA and infectious virus in organs and tissues reflected its tissue specificity.
Fig. 1. Study design. The infection dose of 4.0 log10 TCID50 per animal.
SARS-CoV-2 RNA quantification
The accumulation of viral RNA in organs and tissues was assessed by quantitative reverse transcription polymerase chain reaction as described previously [23]. Viral RNA was isolated from samples using the MagnoPrime UNI reagent kit (NextBio). To detect viral RNA, primers and probe designed for the SARS-CoV-2 nucleocapsid (N) gene were used, as proposed by J. Chan et al. [24].
Histologic examination of the lungs
The right hamster lung was fixed in 10% neutral buffered formalin (BioVitrum) for 24 h, dehydrated according to the standard histological technique and placed in Histomix paraffin medium (BioVitrum). On the Leica RM 2125 RTS rotary microtome we made stepwise longitudinal sections 3-5 microns thick, the preparations were stained with hematoxylin and eosin, enclosed in Canadian balsam (Sigma-Aldrich). Histological preparations were examined using a BX51 light microscope (Olympus). Photofixation of the obtained lung histologic preparations was performed with the help of an Olympus XC10 camera. Pathomorphologic changes in the lungs were evaluated by 2 specialists using a blind method, using a combined severity score from 0 to 3 for each of the morphologic criteria proposed by A.D. Gruber et al. [25]. The maximum possible score was 60.
Statistical processing of data
Statistical analysis was performed using the Graphpad Prism v. 8.0.01 software. Data are presented in graphs as mean, standard deviation (SD), standard error (SE), median, upper and lower quartiles. In box plots, the boundaries of the box are the upper and lower quartiles of the sample (25% and 75%), the ends of the whiskers are the boundaries of a statistically significant sample (without outliers), the line in the box itself is the median of the data. Statistical processing of the obtained results was carried out using the nonparametric method (Mann-Whitney U-test). Differences were considered statistically significant at p < 0.05.
Results
Morphologic changes were absent in histologic preparations of the right lung of uninfected animals (Fig. 2).
Fig. 2. Bronchointerstitial pneumonia in hamster on the 4th day post-challenge with different SARS-CoV-2 strains.
On the 4th day after infection, broncho-interstitial pneumonia was detected in histologic preparations of hamster lungs of all groups (Fig. 2). However, there were significant differences in the severity and prevalence of alterative-inflammatory changes between the groups.
On the 4th day after infection, the groups of animals infected with Wuhan-like Dubrovka strain and FEB2 strain (BA 5.2) showed similar in nature and severity inflammatory changes, the morphological picture of which corresponded to bronchointerstitial pneumonia in the viral stage. The lumen of bronchi and bronchioles in the foci of pneumonia often contained cellular debris, macrophages and neutrophils. The integrity of epithelial lining was focally disturbed due to migration of lymphoid cells, dystrophy, necrosis and desquamation of epitheliocytes. There were loci of epithelial hyperplasia. The wall of bronchi and bronchioles was moderately infiltrated with lymphocytes, histiocytes with a small admixture of polymorphonuclear lymphocytes. Dilated lymphatic vessels located along the course of the bronchial tree contained clusters of lymphocytes. Large lymphoid accumulations (hyperplasia of bronchoassociated lymphoid tissue) were found in the bronchial bifurcation zones. Inflammatory changes were also observed in the walls of medium and small branches of the pulmonary artery accompanying the airways. Perivascular lymphoid tissue was in a state of sharp hyperplasia.
Large confluent foci of pneumonia were observed in all lobes of the organ and were located along the course of the bronchial tree, spreading to the periphery. Their area, estimated at qualitative level, occupied 50-90% of the histologic section area of the organ. Respiratory department in the pneumonia foci represented airless fields, lumen of alveoli in which were not defined, interalveolar septa were destroyed due to expressed lymphoid-histiocytic infiltrate with insignificant admixture of neutrophils. Remains of dead cells nuclei, fibroblasts, erythrocytes were seen among the cells of inflammatory infiltrate. In fresher areas of pneumonia airiness of respiratory section was reduced due to sharp thickening of interalveolar septa and expressed exudation into the alveolar cavity of liquid blood and cells of inflammatory infiltrate: macrophages, lymphocytes, erythrocytes. Many alveoli contained eosinophilic filamentous material (presumably fibrin). In the interalveolar septa there was microvascular hypertension, interstitial edema and diffusely scattered lymphoid-histiocytic infiltrate.
On the 4th day after infection with Podolsk (Delta) strain, the severity and prevalence of inflammatory changes in hamster lungs were lower compared to those in the groups of animals infected with Wuhan-like virus and FEB2 strain (BA.5.2). Small foci of interstitial pneumonia were not located in all lobes, were located along the course of large lobular and segmental bronchi, and their area did not exceed 50% of the histologic section area of the organ. The lumen of bronchi and bronchioles in the foci of pneumonia were mostly free, contained single macrophages, lymphocytes, small groups of desquamated epitheliocytes. The epithelial lining looked preserved over a large length, with single lymphocytes in the field of view of the ×20 objective lens among the cells of the mesenteric epithelium. Airiness of pulmonary parenchyma in the foci of pneumonia was reduced due to thickening of interalveolar septa. Small groups of macrophages, lymphocytes, single neutrophils, erythrocytes and few dead cells (presumably, alveolocytes) were observed in the alveolar cavity. Proteinaceous exudate in the lumen of the alveoli was rare. At this period of the experiment airless and confluent foci of pneumonia were practically absent.
In histologic preparations of hamster lungs euthanized on the 4th day after infection with Otradnoe strain (BA.1.1), the least pronounced pathomorphological changes were observed compared to other groups. Small foci of interstitial pneumonia, which occupied no more than 5–7% of the total section area, were located in 2–3 lobes mainly in the root areas along the course of lobular bronchi. Inflammatory changes in the wall of bronchi and accompanying vessels were weakly expressed.
During histologic examination of the lungs of infected and uninfected hamsters, the morphologic manifestations of coronavirus pneumonia were graded using the recommendations of A.D. Gruber et al. [25]. In infected animals, the cumulative score reflecting the severity of the inflammatory process ranged from 20.8 to 49.8, while in uninfected animals it was close to zero (Fig. 3). In the group of animals infected with Wuhan-like virus, the mean value of the cumulative severity score was 50 ± 6, Delta — 30 ± 5, BA.1.1 — 21 ± 7, BA.5.2 — 39 ± 6.
Fig. 3. Histopathology score of hamster lungs on day 4 post-infection with different SARS-CoV-2 strains. *р < 0.05; **р < 0.01.
In addition to the severity of pathologic changes in hamster lungs, weight dynamics was an important criterion in assessing the virulence of different SARS-CoV-2 strains. The greatest difference in weight of infected and uninfected animals was observed on the 3rd or 4th day after infection. In the group of animals infected with Wuhan-like virus, the delay in weight gain was 14.6% compared to uninfected animals. The similar figure in animals infected with Delta, BA.1.1 and BA.5.2 averaged 2–3% (Fig. 4).
Fig. 4. Weight dynamics in hamsters infected intranasally with different SARS-CoV-2 strains. K– — uninfected hamsters.
Since the main target organs for SARS-CoV-2 are the lungs, nasal passages and brain, not only the viral RNA content but also the infectious activity of the virus was investigated in these organs. The mean values of virus titer in tissues and organs of animals differed significantly depending on the strain used for infection. Thus, on the 4th day after infection, the highest titer values were observed in lung homogenates in groups of animals infected with Delta and Wuhan-like viruses — on average 7.4 log10 and 6.2 log10 TCID50/mL of homogenate, whereas in groups infected with BA.1.1 and BA.5.2, the virus titer was significantly lower — 4.6 and 5.0 log10 TCID50/mL of homogenate, respectively (Fig. 5). In nasal passages homogenates, infectious virus was detected in animals of all groups at a titer of 4.9–6.8 log10 TCID50/mL of homogenate. In brain tissue, infectious virus was detected only in animals infected with Wuhan-like virus (on average 3.7 log10 TCID50/mL of homogenate). It should be noted that the tissue homogenates were toxic to the Vero cells in which the titration was performed; therefore, the limit of sensitivity was 2.0 log10 TCID50/mL of homogenate.
Fig. 5. Titer values of different SARS-CoV-2 strains in the organs of Syrian hamsters on the 4th day after infection. *р < 0.05; **р < 0.01.
In the lungs of infected animals, the concentration of viral RNA varied depending on the strain from 7.6 to 9.3 on average, in the nasal passages from 8.3 to 9.3, and in the brain from 3.8 to 7.6 log10 RNA copies/mL of homogenate (Fig. 6). The highest level of viral RNA in the lungs, nasal passages, brain and other organs of hamsters was observed in the groups of animals infected with Wuhan-like virus and Delta. The concentration of viral RNA in brain homogenates of animals infected with Wuhan-like virus was 7.6, Delta — 5.6, BA.1.1 and BA.5.2 — 3.8 and 4.1 log10 RNA copies/mL, respectively.
Fig. 6. Distribution of viral RNA in the organs of hamsters infected with different SARS-CoV-2 strains. *р < 0.05; **р < 0.01.
Viral RNA was also detected in the heart, liver, kidney, spleen, and blood of most infected animals, but at much lower levels than in the lungs and nasal passages (Fig. 6). The concentration of viral RNA in the above organs of animals infected with BA.1.1 and BA.5.2 was significantly (p < 0.05) lower than in Wuhan-like virus and Delta infection (Fig. 6). The lowest viral RNA content in organs was observed in BA.5.2 infection, while no viral RNA was detected in the blood, kidney and spleen of most animals. It is noteworthy that in all groups of infected animals a significant content of viral RNA was observed in the heart — from 4.7 to 6.1 log10 RNA copies/mL of homogenate.
Discussion
The design of the study implied equality of all conditions, including a single dose of 104 TCID50/head, except that different virus strains were administered to animals of different groups. Since SARS-CoV-2 is a respiratory virus and is transmitted by airborne droplets, the correctness of our choice of intranasal route of virus administration in COVID-19 modeling is undoubted. This method of administration mimics the natural route of infection and is the simplest, fastest and non-invasive way to infect small laboratory animals such as mice and hamsters [4].
Our study revealed differences in virulence and tissue specificity of SARS-CoV-2 strains belonging to different variants of concern. The greatest virulence was possessed by the Wuhan-like Dubrovka strain, which was manifested by the development of subtotal pneumonia and maximum weight gain delay by 14.6% on average. Hamsters infected with Podolsk (Delta), Otradnoe and FEB2 strains (Omicron BA.1 and BA.5.2) lost significantly less weight, 2-3% (p > 0.05). Greater weight loss and severe pneumonia in hamsters infected with Wuhan-like virus was associated with increased virus content in organs and viral damage to the brain. The neurovirulence of Wuhan-like virus was manifested by significantly higher viral RNA content in the brain and isolation of infectious virus from brain homogenates. A number of studies [26, 27] revealed different tropism of SARS-CoV-2 variants to brain cells and lower neurovirulence of the Omicron variant compared to Wuhan-like virus and Alpha and Delta variants, which were dominant earlier [26, 27]. Comparison of the literature data with our own data on the increased tropism of Wuhan-like virus to brain tissues suggests that it was the lesion of the central nervous system that could negatively affect the weight and general condition of the animals [28].
The results of histological examination of the lungs confirmed the data on the different virulence of the virus strains used to infect hamsters. Wuhan-like virus caused the most severe lesions in the lungs with extensive foci of bronchointerstitial pneumonia (cumulative severity score of 50) than Delta- and Omicron-like viruses (cumulative severity score of 21 to 39); p < 0.05. The results obtained, indicating lower virulence for hamsters of Omicron-like strains compared to the ancestral Wuhan virus, are consistent with the lower pathogenicity of the Omicron variant for humans [29], as confirmed by the lower reproductive activity of the virus in human Calu-3 lung cell culture [30]. On the other hand, early conclusions about the lower virulence of Omicron-like strains may have overestimated their attenuation for humans, since they did not separate the real decrease in virulence from the effect of prior immunity, since vaccinated and re-infected individuals naturally carry the disease more easily.
The severity of pathological changes in the lungs during infection with different strains of Omicron and Delta variants also differed significantly: the mean value of the cumulative severity score was 21 ± 7 for BA.1.1, 39 ± 6 for BA.5.2, and 30 ± 5 for Delta. The observed higher virulence of BA.5.2 compared to Delta (p < 0.05) does not agree with the data presented in the article by S. Mohandas et al. [31], who found greater virulence of the Delta-like strain compared to the Omicron variant BA.5.2 sublineage. In this connection, it is important to note that the virulence of different virus strains may be determined not only by their belonging to a particular genetic variant, but also by strain-specific differences and the number of passages the virus isolate underwent in cell culture. It is known that virus isolation and its passages in cell culture are accompanied by the accumulation of mutations that promote virus adaptation to a new host, while virulence decreases for model laboratory animals [32]. Thus, the greater virulence of strain FEB2 (BA.5.2) can be explained by the fact that in our study this strain had undergone 4 passages in Vero cell culture before infection of hamsters, whereas the Podolsk (Delta) strain underwent 16 passages and the Otradnoe (BA.1.1) strain underwent 8 passages.
It is noteworthy that a significant content of viral RNA (up to 6.1 log10 RNA copies/mL) was detected in the heart of animals infected with different strains of SARS-CoV-2. Heart damage by SARS-CoV-2 virus in hamsters has been observed in a number of studies [33, 34]. This observation is also interesting in the context of the high probability of myocarditis in humans after COVID-19. Since ACE2 receptor expression is upregulated in human myocytes [35], the probability of SARS-CoV-2 virus infection of cardiac tissues and the risk of myocarditis development are increased [36, 37].
Among small laboratory animals, COVID-19 modeling is possible in various mouse lines, with the K18-hACE2 transgenic mice being the most susceptible to SARS-CoV-2 [5]. However, at the moment this line of mice is difficult to access, and effective reproduction of the virus in organs other than lungs makes it difficult to use this animal model for modeling viral pneumonia, because these animals have a high percentage of mortality due to causes unrelated to pneumonia. Thus, the main cause of death in K18-hACE2 mice is central nervous system damage and development of viral encephalitis and other neurological diseases due to high expression of ACE2 receptor in brain cells [18, 38, 39]. The disease in K18-hACE2 mice is more severe and has differences in the character of clinical manifestations compared to those in humans.
In view of the above, the hamster model for coronavirus pneumonia is one of the most adequate, accessible and informative among small laboratory animals. Hamsters, when infected with SARS-CoV-2, show clinical signs of respiratory disease and develop mild to moderate pneumonia [18, 40]. Furthermore, they have the ability to spread the virus with infection of contact naive animals [3, 39]. The hamster-based animal model has been widely used in preclinical trials of antiviral drugs and vaccines because it reproduces the development of viral pneumonia without animal death [13, 18, 41, 42]. This study showed that the modeling of COVID-19 caused by new virus variants (Delta, BA.1.1 and BA.5.2) in hamsters remains relevant. These virus variants retained the ability to cause pneumonia with extensive lesions in hamsters. While weight dynamics as an indicator of virulence has become less informative, such indicators as viral load (virus infectivity and viral RNA content in organs) and severity of inflammatory changes in the lungs have retained their informative value in assessing the severity of the disease. Scoring of the severity of pathomorphologic changes in the lungs is of particular value in comparing the severity of pneumonia, because it reduces subjectivity in the evaluation of the results of histologic examination and gives a semi-quantitative assessment of the pathologic process.
Conclusion
The results of this study showed that infection in Syrian golden hamsters infected with SARS-CoV-2 strains belonging to different evolutionary lineages proceeds differently. The Wuhan-like virus was found to be more virulent and neurotropic than the Delta and Omicron variants, which became widespread later. Modeling on hamsters of COVID-19 caused by sublineages of the Omicron variant remains relevant, despite insignificant weight loss of animals, in contrast to infection with Wuhan-like virus. Histologic examination and such indicators as viral load in the lungs, nasal passages, brain, heart and a number of other organs continue to play a decisive role in assessing the pathogenicity of Omicron-like strains for hamsters.
About the authors
Anastasiia V. Gracheva
I.I. Mechnikov Research Institute for Vaccines and Sera
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0001-8428-4482
researcher, Applied virology laboratory
Russian Federation, MoscowAndrey O. Drokov
I.I. Mechnikov Research Institute for Vaccines and Sera
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0009-0006-3300-8078
student, intern, Applied virology laboratory
Russian Federation, MoscowDaria I. Smirnova
I.I. Mechnikov Research Institute for Vaccines and Sera
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0001-7325-0834
junior researcher, Applied virology laboratory
Russian Federation, MoscowDarya M. Khokhlova
I.I. Mechnikov Research Institute for Vaccines and Sera
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0009-0003-5745-7589
junior researcher, Applied virology laboratory
Russian Federation, MoscowEkaterina R. Korchevaya
I.I. Mechnikov Research Institute for Vaccines and Sera
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0002-6417-3301
junior researcher, Applied virology laboratory
Russian Federation, MoscowAndrey A. Pankratov
P.A. Hertsen Moscow Oncology Research Institute — the branch of the National Medical Research Radiological Center
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0001-7291-9743
Cand. Sci. (Biol.), Head, Department of experimental pharmacology and toxicology
Russian Federation, MoscowGalina V. Trunova
P.A. Hertsen Moscow Oncology Research Institute — the branch of the National Medical Research Radiological Center
Author for correspondence.
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0003-2917-4496
Cand. Sci. (Biol.), senior researcher, Department of experimental pharmacology and toxicology
Russian Federation, MoscowVarvara A. Khokhlova
P.A. Hertsen Moscow Oncology Research Institute — the branch of the National Medical Research Radiological Center
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0002-0339-2068
junior researcher, Department of experimental pharmacology and toxicology
Russian Federation, MoscowMaria S. Vorontsova
P.A. Hertsen Moscow Oncology Research Institute — the branch of the National Medical Research Radiological Center
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0002-9320-1746
Cand. Sci. (Biol.), junior researcher, Department of experimental pharmacology and toxicology
Russian Federation, MoscowIrina A. Leneva
I.I. Mechnikov Research Institute for Vaccines and Sera
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0002-7755-2714
D. Sci. (Biol.), Head, Experimental virology laboratory
Russian Federation, MoscowOksana A. Svitich
I.I. Mechnikov Research Institute for Vaccines and Sera; I.M. Sechenov First Moscow State Medical University (Sechenov University)
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0003-1757-8389
D. Sci. (Med.), Prof., Corresponding Member of RAS, Director, Prof., Department of microbiology, virology and immunology
Russian Federation, Moscow; MoscowVitaly V. Zverev
I.I. Mechnikov Research Institute for Vaccines and Sera; I.M. Sechenov First Moscow State Medical University (Sechenov University)
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0001-5808-2246
D. Sci. (Biol.), Prof., Academician of RAS, scientific director, Head, Department of microbiology, virology and immunology
Russian Federation, Moscow; MoscowEvgeny B. Faizuloev
I.I. Mechnikov Research Institute for Vaccines and Sera; Russian Medical Academy of Continuous Professional Education
Email: anastasiia.gracheva.95@mail.ru
ORCID iD: 0000-0001-7385-5083
Cand. Sci. (Biol.), Head, Applied virology laboratory, senior lecturer, Department of virology
Russian Federation, Moscow; MoscowReferences
- Fan C., Wu Y., Rui X., et al. Animal models for COVID-19: advances, gaps and perspectives. Signal Transduct. Target. Ther. 2022;7(1):220. DOI: https://doi.org/10.1038/s41392-022-01087-8
- Roberts A., Vogel L., Guarner J., et al. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J. Virol. 2005;79(1):503–11. DOI: https://doi.org/10.1128/jvi.79.1.503-511.2005
- Chan J.F., Zhang A.J., Yuan S., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: Implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71(9):2428–46. DOI: https://doi.org/10.1093/cid/ciaa325
- Qi F., Qin C. Characteristics of animal models for COVID-19. Animal Model. Exp. Med. 2022;5(5):401–9. DOI: https://doi.org/10.1002/ame2.12278
- Leneva I.A., Smirnova D.I., Kartashova N.P., et al. Comparative study of Wuhan-like and omicron-like variants of SARS-CoV-2 in experimental animal models. Vopr. Virusol. 2022;67(5):439–49. DOI: https://doi.org/10.36233/0507-4088-135
- Kirk N.M., Liang Y., Ly H. Pathogenesis and virulence of coronavirus disease: Comparative pathology of animal models for COVID-19. Virulence. 2024;15(1):2316438. DOI: https://doi.org/10.1080/21505594.2024.2316438
- Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., et al. Animal models for COVID-19. Nature. 2020;586(7830):509–15. DOI: https://doi.org/10.1038/s41586-020-2787-6
- Fenollar F., Mediannikov O., Maurin M., et al. Mink, SARS-CoV-2, and the human-animal interface. Front. Microbiol. 2021;12:663815. DOI: https://doi.org/10.3389/fmicb.2021.663815
- Kutter J.S., de Meulder D., Bestebroer T.M., et al. SARS-CoV and SARS-CoV-2 are transmitted through the air between ferrets over more than one meter distance. Nat. Commun. 2021;12(1):1653. DOI: https://doi.org/10.1038/s41467-021-21918-6
- Ciurkiewicz M., Armando F., Schreiner T., et al. Ferrets are valuable models for SARS-CoV-2 research. Vet. Pathol. 2022;59(4):661–72. DOI: https://doi.org/10.1177/03009858211071012
- Martins M., Nooruzzaman M., Cunningham J.L., et al. The SARS-CoV-2 spike is a virulence determinant and plays a major role on the attenuated phenotype of Omicron virus in a feline model of infection. J. Virol. 2024;98(3):e0190223. DOI: https://doi.org/10.1128/jvi.01902-23
- Bao L., Deng W., Huang B., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583(7818):830–3. DOI: https://doi.org/10.1038/s41586-020-2312-y
- Xue Y., Yang D., Vogel P., et al. Cardiopulmonary Injury in the Syrian hamster model of COVID-19. Viruses. 2022;14(7):1403. DOI: https://doi.org/10.3390/v14071403
- Miao J., Chard L.S., Wang Z., Wang Y. Syrian hamster as an animal model for the study on infectious diseases. Front. Immunol. 2019;10:2329. DOI: https://doi.org/10.3389/fimmu.2019.02329
- Iwatsuki-Horimoto K., Nakajima N., Ichiko Y., et al. Syrian hamster as an animal model for the study of human influenza virus infection. J. Virol. 2018;92(4):e01693–17. DOI: https://doi.org/10.1128/JVI.01693-17
- Fan S., Gu C., Kong H., et al. Influenza viruses suitable for studies in Syrian hamsters. Viruses. 2022;14(8):1629. DOI: https://doi.org/10.3390/v14081629
- Toth K., Lee S.R., Ying B., et al. STAT2 knockout Syrian hamsters support enhanced replication and pathogenicity of human adenovirus, revealing an important role of type I interferon response in viral control. PLoS Pathog. 2015;11(8):e1005084. DOI: https://doi.org/10.1371/journal.ppat.1005084
- Rosenke K., Meade-White K., Letko M., et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020;9(1):2673–84. DOI: https://doi.org/10.1080/22221751.2020.1858177
- Xu J., Liu M., Niu X., et al. The cold-adapted, temperature-sensitive SARS-CoV-2 strain TS11 is attenuated in Syrian hamsters and a candidate attenuated vaccine. Viruses. 2022;15(1):95. DOI: https://doi.org/10.3390/v15010095
- Wang Y., Yang C., Song Y., et al. Scalable live-attenuated SARS-CoV-2 vaccine candidate demonstrates preclinical safety and efficacy. Proc. Natl. Acad. Sci. U.S.A. 2021;118(29):e2102775118. DOI: https://doi.org/10.1073/pnas.2102775118
- Faizuloev E.B., Gracheva A., Korchevaya E.R., et al. Single intranasal immunization with attenuated Wuhan-like SARS-CoV-2 provides highly effective cross-protection against Delta and Omicron variants of concern: 1. J. Microbiol. Epidemiol. Immunobiol. 2024;101(1):36–51. DOI: https://doi.org/10.21203/rs.3.rs-3279049/v1
- Ramakrishnan M.A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 2016;5(2):85–6. DOI: https://doi.org/10.5501/wjv.v5.i2.85
- Gracheva A.V., Korchevaya E.R., Ammour Y.I., et al. Immunogenic properties of SARS-CoV-2 inactivated by ultraviolet light. Arch. Virol. 2022;167(11):2181–91. DOI: https://doi.org/10.1007/s00705-022-05530-7
- Chan J.F., Yip C.C., To K.K., et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e00310-20. DOI: https://doi.org/10.1128/JCM.00310-20
- Gruber A.D., Osterrieder N., Bertzbach L.D., et al. Standardization of reporting criteria for lung pathology in SARS-CoV-2-infected hamsters: what matters? Am. J. Respir. Cell Mol. Biol. 2020;63(6):856–9. DOI: https://doi.org/10.1165/rcmb.2020-0280LE
- Bauer L., van Riel D. Do SARS-CoV-2 variants differ in their neuropathogenicity? mBio. 2023;14(1):e0292022. DOI: https://doi.org/10.1128/mbio.02920-22
- Bauer L., Rissmann M., Benavides F.F.W., et al. In vitro and in vivo differences in neurovirulence between D614G, Delta and Omicron BA.1 SARS-CoV-2 variants. Acta Neuropathol. Commun. 2022;10(1):124. DOI: https://doi.org/10.1186/s40478-022-01426-4
- Bauer L., Laksono B.M., de Vrij F.M.S., et al. The neuroinvasiveness, neurotropism, and neurovirulence of SARS-CoV-2. Trends Neurosci. 2022;45(5):358–68. DOI: https://doi.org/10.1016/j.tins.2022.02.006
- Trunfio M., Portesani F., Vicinanza S., et al. Real-life evidence of lower lung virulence in COVID-19 inpatients infected with SARS-CoV-2 Omicron variant compared to wild-type and Delta SARS-CoV-2 pneumonia. Lung. 2022;200(5):573–7. DOI: https://doi.org/10.1007/s00408-022-00566-7
- Purwono P.B., Vacharathit V., Manopwisedjaroen S., et al. Infection kinetics, syncytia formation, and inflammatory biomarkers as predictive indicators for the pathogenicity of SARS-CoV-2 Variants of Concern in Calu-3 cells. PLoS One. 2024;19(4):e0301330. DOI: https://doi.org/10.1371/journal.pone.0301330
- Mohandas S., Shete A., Kumar A., et al. Comparative pathogenicity of BA.2.12, BA.5.2 and XBB.1 with the Delta variant in Syrian hamsters. Front. Microbiol. 2023;14:1183763. DOI: https://doi.org/10.3389/fmicb.2023.1183763
- Li X.F., Cui Z., Fan H., et al. A highly immunogenic live-attenuated vaccine candidate prevents SARS-CoV-2 infection and transmission in hamsters. Innovation (Camb). 2022;3(2):100221. DOI: https://doi.org/10.1016/j.xinn.2022.100221
- Daems M., Liesenborghs L., Boudewijns R., et al. SARS-CoV-2 infection causes prolonged cardiomyocyte swelling and inhibition of HIF1α translocation in an animal model COVID-19. Front. Cardiovasc. Med. 2022;9:964512. DOI: https://doi.org/10.3389/fcvm.2022.964512
- Jones E.A.V. Mechanism of COVID-19-induced cardiac damage from patient, in vitro and animal studies. Curr. Heart Fail Rep. 2023;20(5):451–60. DOI: https://doi.org/10.1007/s11897-023-00618-w
- Liu H., Gai S., Wang X., et al. Single-cell analysis of SARS-CoV-2 receptor ACE2 and spike protein priming expression of proteases in the human heart. Cardiovasc. Res. 2020;116(10):1733–41. DOI: https://doi.org/10.1093/cvr/cvaa191
- Ishisaka Y., Watanabe A., Aikawa T., et al. Overview of SARS-CoV-2 infection and vaccine associated myocarditis compared to non-COVID-19-associated myocarditis: a systematic review and meta-analysis. Int. J. Cardiol. 2024;395:131401. DOI: https://doi.org/10.1016/j.ijcard.2023.131401
- Thaker R., Faraci J., Derti S., Schiavone J.F. Myocarditis in SARS-CoV-2: A meta-analysis. Cureus. 2023;15(10):e48059. DOI: https://doi.org/10.7759/cureus.48059
- Jiang R.D., Liu M.Q., Chen Y., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182(1):50-58.e8. DOI: https://doi.org/10.1016/j.cell.2020.05.027
- Sia S.F., Yan L.M., Chin A.W.H., et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020;583(7818):834–8. DOI: https://doi.org/10.1038/s41586-020-2342-5
- Imai M., Iwatsuki-Horimoto K., Hatta M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U.S.A. 2020;117(28):16587–95. DOI: https://doi.org/10.1073/pnas.2009799117
- Yuan S., Ye Z.W., Liang R., et al. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science. 2022;377(6604):428–33. DOI: https://doi.org/10.1126/science.abn8939
- Mohandas S., Yadav P.D., Sapkal G., et al. Pathogenicity of SARS-CoV-2 Omicron (R346K) variant in Syrian hamsters and its cross-neutralization with different variants of concern. EBioMedicine. 2022;79:103997. DOI: https://doi.org/10.1016/j.ebiom.2022.103997
Supplementary files