The incidence of chronic viral hepatitis and the analysis of the genetic diversity of hepatitis B and C viruses among the population of Khabarovsk city
- Authors: Kotova V.O.1, Bazykina E.A.1, Balakhontseva L.A.1, Trotsenko O.E.1, Kuznetsova A.V.2
-
Affiliations:
- Khabarovsk Research Institute of Epidemiology and Microbiology
- Center for Prevention and Combat Against AIDS and Infectious Diseases
- Issue: Vol 102, No 1 (2025)
- Pages: 91-101
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18640
- DOI: https://doi.org/10.36233/0372-9311-578
- EDN: https://elibrary.ru/zdffei
- ID: 18640
Cite item
Abstract
Introduction. Evaluation of hepatitis virus genotypes diversity plays a significant role in analysis of epidemiological situation in particular territories which allows to trace possible routs of transmission, emergence and spread of new viral variants.
The aim of the study is to evaluate dynamic of incidence of chronic forms of viral hepatitis B and C in the Khabarovsk krai during 2013-2023 and to perform analysis of genetic diversity of hepatitis B and C viruses (HBV and HCV) that were circulating in the Khabarovsk city.
Materials and methods. A retrospective analysis of the incidence of chronic forms of hepatitis C in the Khabarovsk krai was conducted for the period from 2013 to 2023 (11 years) according to reports from the Office of Rospotrebnadzor in the Khabarovsk krai. Serological and molecular-genetic analysis of 112 blood plasma samples obtained from patients with diagnosis of “chronic viral hepatitis” residing in the Khabarovsk city was conducted.
Results. The research showed that incidence of chronic viral hepatitis C has declined from 50.0 in 2013 to 44.6 cases per 100 thousand population in 2023 and incidence of chronic viral hepatitis B has also declined from 10.2 to 8.2 cases per 100 thousand population in the Khabarovsk krai respectively. HBV DNA was detected in 21 out of 36 blood plasma samples that contained serological markers of HBV. HBV genotype D was isolated in 15 out of 17 samples and was presented by two subtypes (D1, D2). Subtype A2 was identified in 2 samples. No mutations of drug resistance were found. HCV RNA was found in 58 out of 78 samples of blood plasma that were positive for antibodies (IgG + IgM) to HCV. Circulation of 1b, 1a, 3a, 2a, 2c HCV subtypes with predominance of 1b subtype was revealed in the Khabarovsk city.
Conclusion. The incidence of chronic hepatitis B and C in the Khabarovsk krai during the 11-year follow-up period had a pronounced tendency to decrease. Results of the research complement existing data on circulation of HBV and HCV genetic variants in territories of the Russian Federation.
Full Text
Introduction
The problem of parenteral viral hepatitis B and C remains one of the priority objectives of global health care due to the high frequency of chronic forms of the disease and the possibility of unfavorable outcomes in the form of liver cirrhosis and hepatocellular carcinoma. The World Health Organization (WHO) estimates that 50 million people worldwide are affected by hepatitis C virus (HCV) and 254 million people are infected with hepatitis B virus (HBV)1. In April 2016, the WHO Assembly resolved to globally eliminate viral hepatitis as a public health problem by 2030.2
The implementation of the “Health” national project on hepatitis B vaccine prophylaxis in Russia has contributed to a decrease in the incidence of acute form of the infection. Over the past 10 years (from 2014 to 2023), the incidence of acute hepatitis B has decreased 4-fold — from 1.32 to 0.33 cases per 100,000 population. Since 2014, there has been an annual decrease in the incidence of acute hepatitis C in Russia. In 2023, it amounted to 1393 patients or 0.95 cases per 100,000 population, which is 27.5% lower than the global average, but at the same time, compared to 2022, the incidence increased by 26%. Along with a decrease in the incidence of acute hepatitis B and C, high rates of new cases of chronic viral hepatitis continue to be registered in Russia. In total, more than 58.9 thousand cases of chronic viral hepatitis were registered in 2023 (43.3 thousand cases in 2022). The incidence rates of chronic viral hepatitis vary dramatically across the constituent entities of the Russian Federation (from 0.9 to 127.86 per 100,000 population), which largely depends on the quality of diagnosis and completeness of registration of this group of diseases3. A similar trend can be observed in the Khabarovsk Territory, which is part of the Far Eastern Federal District4.
Recently, much attention has been paid to the study of genotypic variability of hepatitis viruses. Diseases caused by different genotypes may differ significantly in clinical course and outcomes [1–3].
Currently, 10 hepatitis B virus (HBV) genotypes have been identified (designated by letters A to J). Genotypes A–D, F, H and I are divided into 35 sub-genotypes (for other genotypes sub-genotypes have not been established) [4, 5]. The genotypes differ in genome length, size of the open reading frames and protein translated, as well as mutations selected under the influence of therapy [6, 7]. Hepatitis C virus (HCV) isolates are categorized into 8 genotypes [8] and 93 confirmed subtypes. The modern classification of HCV includes 9 inter-genotype recombinant forms5. Each genotype is characterized by a certain frequency of occurrence and geographical area of distribution.
According to the results of molecular genetic studies conducted in different years in Russia, it was found that 3 genotypes of HBV — D, A and C with the predominance of genotype D and 4 subtypes of HCV — 1a, 1b, 2 and 3a, of which subtypes 1b and 3a are predominant, are circulating in the territory of the country [9–12].
The study of genotypic diversity of viral hepatitis is of great importance when analyzing the epidemiological situation in each specific territory, allowing to track possible routes of transmission, the emergence and spread of new virus variants, identify imported cases of infection. All this is an important aspect in the timely planning of necessary sanitary and epidemiologic measures.
Modern molecular biological methods of research allow not only to assess the genetic diversity of HBV and HCV in separate territories, but also to determine the nature of clinically significant mutations in the P-gene of HBV DNA and NS3/NS5A/NS5B regions of HCV associated with possible resistance to therapy with direct antiviral drugs.
The aim of the study was to assess the dynamics of chronic hepatitis B and C incidence in 2013–2023, and to analyze the genetic diversity of HBV and HCV circulating among Khabarovsk patients diagnosed with chronic viral hepatitis.
Materials and methods
Retrospective analysis of the incidence of chronic hepatitis B and C was performed using data provided by the Khabarovsk krai Rospotrebnadzor regional office for the period from 2013 to 2023 (11 years). The values of chronic viral hepatitis incidence in Russia were taken from the State Report “On the state of sanitary and epidemiologic well-being of the population in the Russian Federation in 2023”.
The average annual growth rate (AAGR) was calculated using the least squares method, and the confidence interval was calculated using the Fisher angular transformation method. The obtained trend was regarded as stable at AAGR = ± 1%, as weakly expressed —at AAGR = ± 1.1–3.0%, as moderate – at AAGR = ± 3.1–5.0%, as expressed – at AAGR = ± 5.1–7.0%, as pronounced – at AAGR = ±7% and above [13].
In order to study the genetic diversity of HBV and HCV circulating among the population of the Khabarovsk Krai, blood plasma samples from 112 patients diagnosed with chronic viral hepatitis collected at the Center for Prevention and Control of AIDS and Infectious Diseases (n = 50) and the laboratory of the Khabarovsk Research Institute of Epidemiology and Microbiology (n = 62) in 2022–2023 were studied. Informed consent of all patients to participate in the study was obtained, as well as approval of the ethics committee of Khabarovsk Research Institute of Epidemiology and Microbiology (protocol No. 9 of 01.11.2022) to conduct the research. The presence of markers of viral hepatitis B and C was determined by enzyme immunoassay using test systems produced by Vector-Best CJSC.
Nucleic acids were isolated from 100 µl of blood plasma using the AmpliPrime RIBO-prep reagent kit (Central Research Institute of Epidemiology of Rospotrebnadzor). Analysis for detection of HBV DNA, HCV RNA, determination of viral load in positive samples and HCV genotype was performed by polymerase chain reaction (PCR) with hybridization-fluorescence detection in real time using commercial kits: “AmpliSens HBV-FL, AmpliSens HBV-Monitor-Fl, AmpliSens HCV-FL, AmpliSens HCV-Monitor-Fl, AmpliSens-1/2/3 (Central Research Institute of Epidemiology of Rospotrebnadzor) on a Rotor Gene Q device (Qiagen).
For HBV and HCV genotyping, we used two-step PCR with specific primers (Syntol) for the conserved region of the overlapping S and P genes, encoding the surface protein and DNA polymerase of HBV, and for the NS5B region of HCV [14, 15].
Additionally, the nucleotide sequence of the HCV NS5 region was analyzed for a patient with HCV receiving direct antiviral drugs using the method described by M. Rajhi et al. [16].
Sanger sequencing of amplified viral genome fragments was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems/Life Technologies) on an ABI3500 DNA analyzer (Applied Biosystems/Life Technologies).
Alignment of the obtained nucleotide sequences was performed in the BioEdit v. 7.1.9 program.
Phylogenetic analysis was performed using the MEGA v. 7.0 program by constructing phylogenetic trees using the neighbor-joining method [17]. The distances between nucleotides were calculated using the Kimura method. To assess the statistical reliability of phylogenetic relationships, bootstrap analysis was used for 1000 independent constructions of each phylogenetic tree.
As reference strains for phylogenetic analysis we used sequences of the same genomic region of HBV and HCV from Russia and other countries of the world presented in GenBank6.
The geno2pheno software7 was used to detect drug resistance mutations.
14 obtained and analyzed nucleotide sequences of HBV genomic regions (accession numbers PP100729-PP100742) and 44 HCV genomic regions (accession numbers PP111728-PP111748, PP100743-PP100765) were deposited in the international GenBank database. 10 HBV partial genomic sequences and 35 HCV partial genomic sequences were deposited in Russian virus genome information aggregation platform VGARus.
Results
The conducted studies showed that the incidence of chronic hepatitis C in the Khabarovsk Krai in 2013 to 2023 decreased from 50.0 (95% CI 46.2–54.0) to 44.6 (95% CI 41.0–48.3) per 100,000 population and had a pronounced downward trend (AAGR = –6.5%). More significant changes were registered in the Khabarovsk Krai in terms of chronic hepatitis B incidence, which had a pronounced downward trend (AAGR = –11.6%), decreasing more than 2-fold — from 10.2 (95% CI 8.5–12.0) cases per 100,000 population in 2013 to 4.5 (95% CI 3.4–5.7) cases per 100,000 population in 2021.
In Khabarovsk, the incidence of both chronic hepatitis B and C was higher than in the Khabarovsk Krai throughout the entire observation period: in 2023 the incidence of chronic hepatitis C was 56.7 (95% CI 50.9–62.8) per 100,000 population (27.1% higher than in the Khabarovsk Krai), and the incidence for chronic hepatitis B was 13.3 (95% CI 10.6–16.3) per 100,000 population (62.2% higher than in the Khabarovsk Krai). In multiyear dynamics, the incidence of chronic hepatitis B and C in Khabarovsk had a more pronounced downward trend compared to the regional one (AAGR = –11.6% — for chronic hepatitis C incidence and AAGR = –12.2% — for chronic hepatitis B incidence) (Fig. 1).
Fig. 1. Long-term incidence of chronic viral hepatitis C and B in the Khabarovsk krai and Khabarovsk city.
The decline in chronic viral hepatitis incidence after 2019 coincided with the start of the COVID-19 pandemic, during which restrictive measures were introduced to reduce the spread of SARS-CoV-2 among the population. In 2022 and 2023, when the restrictions were gradually lifted, an increase in chronic hepatitis C incidence of 20.5 and 45.7% in the Khabarovsk Krai and 21.4 and 44.6% in Khabarovsk, respectively, began to be recorded. The incidence of chronic hepatitis B in the Khabarovsk Krai and Khabarovsk increased by 17.8 and 41.2% in 2022, respectively, and by 54.7 and 84.7% in 2023, which is probably due to an increase in the number of patient visits to medical organizations, including laboratory testing that reached levels of 2019.
Serologic and molecular genetic analysis was performed for 112 blood plasma samples of the Khabarovsk residents diagnosed with chronic viral hepatitis. The age of the examined patients averaged 51 years. There were 51 women (45.5%; 95% CI 36.9–55.2%) and 61 men (54.5%; 95% CI 45.8–64.1%) among the examined patients.
HBsAg was detected in 36 of 112 plasma samples (32.1%; 95% CI 23.7–40.9%). Antibodies to the HCV were detected in 78 samples (69.6%; 95% CI 61.2–78.1). 2 (1.8%; 95% CI 0.2–5.4%) patients had mixed-infection with HBV + HCV.
HBV DNA was detected in 21 (58.3%; 95% CI 41.7–73.4%) of 36 plasma samples from patients with detectable markers of HBV. In all DNA-positive samples, viral load levels were determined. In 19 (90.5%; 95% CI 74.0–98.8%) patients it was low (less than 104 IU/mL), and in 2 (9.5%; 95% CI 1.2–26.0) it was intermediate (104–106 IU/mL).
During an examination of 78 patients from Khabarovsk who were positive by enzyme-linked immunosorbent assay for antibodies (IgG + IgM) to HCV, HCV RNA was detected in 54 (69.2%; 95% CI 42.9–64.9%) blood plasma samples. In all RNA-positive samples, viral load levels were determined. It was low (less than 8 × 105 IU/mL) in 39 (72.2%; 95% CI 59.4–83.1) patients and high (greater than 8 × 105 IU/mL) in 15 (27.8%; 95% CI 16.9–40.6) patients. Preliminary HCV genotyping using the AmpliSens -1/2/3 kit showed that HCV genotype 1 was the most common among the examined patients in Khabarovsk (57.4%; 95% CI 43.7–69.8%). HCV genotype 3 was detected in 14 patients (25,9%; 95% CI 15,3–38,4%). Genotype 2 was detected in 8 cases (14.8%; 95% CI 6.8–25.6%), and genotype could not be determined in 1 patient (1.9%; 95% CI 0.1–7.2%).
To determine the genotype, to clarify the origin and possible relatedness of HBV and HCV variants with isolates from other regions of Russia and foreign countries both nearby and far away, 17 nucleotide sequences of the HBV genomic region and 53 nucleotide sequences of the NS5B region of HCV of satisfactory quality were obtained, suitable for further analysis.
As a result of molecular genetic analysis of HBV, genotype D was determined in 15 patients (88.2%; 95% CI 68.9–98.7%) and genotype A in 2 patients (11.8%; 95% CI 1.3–31.1). Phylogenetic relationships between the studied samples and reference sequences are presented in Fig. 2.
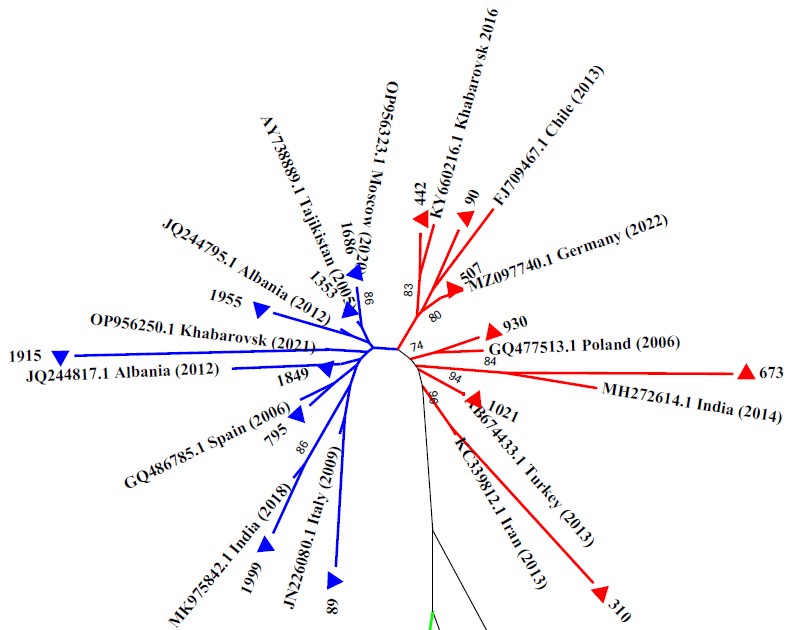
Fig. 2. Results of phylogenetic analysis of partial genomic nucleotide sequences of HBV circulating among population of the Khabarovsk city.
Phylogenetic tree was constructed using the neighbor-joining method. HBV sequences evaluated in current work are marked with triangles. HBV reference sequences are specified by their GenBank accession numbers. Bootstrap index values exceeding 70% are indicated. Blue lines — sub-genotype D2; red lines — sub-genotype D1; green lines — sub-genotype A2.
On the phylogram, the HBV genotype D strains obtained by us and the nucleotide sequences retrieved from GenBank fell into two distinct monophyletic groups distinguished by sub-genotypes: D1 and D2, which were identified in 7 (46.7%) and 8 (53.3%) samples, respectively.
The obtained nucleotide sequences (Nos. 442, 507, 90) grouped with genetically similar sequences of sub-genotype D1 from Chile (FJ709467.1), Germany (MZ0977440.1), as well as with a strain (KY660216.1) that had already been recorded in the Khabarovsk Krai in 2016. Four samples (Nos. 930, 673, 1021, 310) formed separate clades with sub-genotype D1 strains from Poland (GQ477513.1), India (MH272614.1), Turkey (AB674433.1) and Iran (KS339812.1).
The obtained nucleotide sequences of sub-genotype D2 were distributed between HBV strains of this sub-genotype detected in Russia (in Moscow — OR956323. 1 and Khabarovsk — OR956250.1), and strains from India (MK975842.1), Italy (JN226080.1), Spain (GQ486785.1), Albania (JQ244817.1, JQ244795.1), Iran (KC339865.1), Tajikistan (AY738889).
Two strains isolated in Khabarovsk (samples No. 1125, 1877) grouped on the phylogenetic tree, forming one common cluster with reference strains of sub-genotype A2. Sample No. 1877 had a high genetic similarity with a strain of HBV sub-genotype A2, serotype adw2 from Cuba (KM606811.1), and sample No. 1125 with a strain from Poland (GQ477462.1).
The nucleotide sequences of the P-gene region of HBV obtained in the present study were tested for the presence of key mutations associated with the emergence of drug resistance. The analysis showed that all 17 HBV strains were sensitive to the following antiviral drugs: lamivudine, adefovir, entecavir, telbivudine, tenofovir, i.e. no resistance mutations were detected.
Phylogenetic relationships between nucleotide sequences obtained by direct sequencing of the NS5B region of the HCV genome and reference sequences are displayed in Fig. 3.
Fig. 3. Result of phylogenetic analysis of NS5B region nucleotide sequences of HCV circulating among population of the Khabarovsk city.
Phylogenetic tree was constructed using the neighbor-joining method. HCV sequences evaluated in current work are marked with triangles. HCV reference sequences are specified by their GenBank accession numbers. Bootstrap index values exceeding 70% are indicated. Blue lines — subtype 3a; red lines — subtype 1b; green lines — subtype 2a; black lines — subtype 1a; violet lines — subtype 2c.
Sample No. 57, for which no result was obtained during preliminary genotyping using AmpliSens -1/2/3 diagnostic test system, formed a single cluster with isolates belonging to subtype 1a and isolated in different years in the USA (KT734609. 1, OK392383), where this genetic variant is endemic, as well as in Sweden (MH510449.1), Ukraine (OQ979420), Switzerland (EU255927.1), and the Netherlands (KU563369.1), but no closely related strains were identified for it.
On the phylogenetic tree, 31 out of 54 strains under study were evenly distributed among the reference sequences of HCV subtype 1b presented in the international GenBank database (Greece, France, USA, Belgium, Spain, Taiwan, Japan, Vietnam, Brazil, Indonesia, Tunisia). Phylogenetic analysis of 13 specimens assigned to genotype 3 by PCR-genotyping showed that all nucleotide sequences obtained by us are clustered on the same branch of the phylogenetic tree with previously obtained sequences of the same genomic region of variants of subtype 3a isolated in different years in different regions of Russia and the world.
Phylogenetic analysis of 8 specimens assigned to genotype 2 based on the results of preliminary genotyping revealed the formation of 2 clusters. One sample (No. 81) with a high level of bootstrap support (100%) formed a single cluster with isolates belonging to subtype 2c, but no closely related strains were identified for it. Seven strains grouped on the phylogenetic tree, forming one common cluster with HCV subtype 2a isolates registered in GenBank, which were isolated in Russia — in the Novosibirsk region in 2002 (GQ388000.1), Republic of Sakha (Yakutia) (KT 378626.1) in 2014, Leningrad region (AF388438.1) in 2014, and in France (MG453401.1) in 2014.
For sample #76 from a patient with HCV receiving direct antiviral drugs, the nucleotide sequence of the HCV NS5A region was additionally analyzed for the presence of mutations associated with the emergence of drug resistance. As a result of the study, the Y93H mutation was detected, which causes resistance of the virus to the following drugs: Daclatasvir, which was included in the patient's treatment regimen, as well as to Elbasvir, Ledipasvir, Ombitasvir and Velpatasvir. When analyzing the nucleotide sequence of the NS5B region, no drug resistance mutations were detected in this sample.
Discussion
The current epidemiological situation on viral hepatitis both in the Khabarovsk Krai and on average in Russia is characterized by an increase in the total number of patients with chronic forms among the population. In general, in the Khabarovsk Krai for 2013-2023, the curves of chronic hepatitis B and C incidence had unidirectional downward trends. Despite this, the incidence rate of chronic hepatitis C in the Khabarovsk Krai, which reached 44.6 cases per 100,000 population in 2023, exceeded the national average of 31.6 cases per 100,000 population. At the same time, the incidence rate of chronic hepatitis B in the Khabarovsk Krai was slightly lower than the Russian average — 8.2 and 8.4 cases per 100,000 population, respectively8.
It should be noted that the sharp decline in the incidence of chronic hepatitis B and C, recorded in 2020, including for chronic hepatitis C — 1.7-fold from 43.0 (95% CI 39.5–46.6) to 24.8 (95% CI 22.2–27.8) and for chronic hepatitis B — almost 2-fold from 9.8 (95% 8.2–11.6) to 5.0 (95% CI 3.8–6.3) cases per 100,000 population, is partly explained by a decrease in the number of people seeking medical care during the COVID-19 pandemic.
For many years, a significant contribution to the development of the epidemic process of chronic viral hepatitis in the region has been made by the largest population center, Khabarovsk, which is the regional capital, where 61.1% (95% CI 57.1–65.1) of patients with chronic hepatitis C and 78.1% (95% CI 69.7–85.4) of patients with chronic hepatitis B of all first-detected cases in the constituent entity lived in 2023. The higher incidence of chronic hepatitis C in the regional capital can be partly attributed to the greater availability of laboratory testing, in contrast to the northeastern territories remote from the regional center, where registration of chronic viral hepatitis incidence is absent or sporadic, such as Polina Osipenko, Nikolaevsky, Okhotsk, Tuguro-Chumikansky and Ayano-Maysky districts.
A molecular genetic study established HBV and HCV genotypes and sub-genotypes circulating among patients with chronic forms of infection in Khabarovsk.
The results of phylogenetic analysis of D1 and D2 sub-genotypes from Khabarovsk and the GenBank database showed the formation of several clusters, which may indicate different origins and independent spread of HBV strains circulating in the study area. The low similarity of the strains from this study with strains from Russia is possibly due to the low proportion of Russian variants of the virus represented in the international GenBank database.
The study revealed some differences in the distribution of HBV and HCV genetic variants noted in this paper among residents of Khabarovsk and the study we conducted in 2017–2018 among the indigenous population of Nanai district of the Khabarovsk Krai [18, 19]. Thus, analysis of the frequency of occurrence of HBV sub-genotypes D1, D2 and D3 showed that among patients in the rural area, sub-genotype D3 was slightly predominant (51.3%; 95% CI 35.8–66.7%), and sub-genotypes D2 and D1 were identified in 46.1% (95% CI 30.9–61.7%) and 2.6% of cases, respectively. Should be noted that in the present study, no cases of infection with sub-genotype D3 were identified among urban patients, which refutes the results of previously described studies, according to which the incidence of sub-genotype D1 decreases from 45% in the European part of Russia to 12% in the Far Eastern region [20]. This can be partly explained by the small sample size of the study. This fact requires further investigation.
The results of the present study showed that HCV subtype 1b was most prevalent among the examined patients in Khabarovsk, detected by phylogenetic analysis in 58.5% of cases, while the prevalence of subtype 3a (45.0%) was recorded in the Nanai district of the Khabarovsk Krai.
Phylogenetic analysis of the NS5B region of HCV performed for 53 investigated blood samples of Khabarovsk residents initially typed in PCR presented the following ratio of subtypes: 1b — 31 (58.5%; 95% CI 45.2–71.4%), 3a — 13 (24.5%; 95% CI 14.0–37.0%), 2a — 7 (13.2%; CI 5.5–23.5%), with subtypes 1a and 2c (1.9%) identified in isolated cases. According to the results of the phylogenetic analysis, isolate No. 57, untyped by conventional PCR, could be attributed to subtype 1a.
In summary, the obtained data on the circulation of HBV and HCV genetic variants among Khabarovsk residents are consistent with the data of other researchers on the predominance of HBV genotype D and HCV subtypes 1b and 3a in Russia.
It should be noted the importance of regular updating of information on genotypes/subtypes of hepatitis viruses detected in the subjects of the Russian Federation in the international (GenBank) and Russian (VGARus) databases. This will significantly expand the opportunities for molecular genetic monitoring for circulating genetic variants of viruses in a particular territory, as well as for analyzing the incidence of these infections in Russia as a whole, which may ultimately strengthen the effectiveness of epidemiologic surveillance of viral hepatitis. Furthermore, thanks to the use of modern molecular biological diagnostic methods and phylogenetic analysis, it has become possible to predict the development of unfavorable trends in the epidemiological situation, as well as to confirm or deny the presence of an epidemiological link between the suspected source of HCV or HBV and the diseased when investigating the fact of intra-family infection, group diseases, cases of nosocomial infection or occupational exposure to viral hepatitis.
Conclusion
Despite the decrease in cases of detection of acute forms of parenteral viral hepatitis in the Khabarovsk Krai, the registration of chronic viral hepatitis remains at a high level. The most unfavorable situation with the incidence of chronic hepatitis B and C in the Khabarovsk Krai is observed in Khabarovsk.
The study of genetic diversity of HBV and HCV viruses in Khabarovsk revealed the circulation of two HBV genotypes: D and A. Among the samples studied, HBV genotype D was detected in 88.2% of cases and is represented by sub-genotypes D1 and D2. Subtype A accounted for 11.8%. Molecular genetic study of HCV circulating in Khabarovsk revealed circulation of subtypes 1b, 1a, 3a, 2a, 2c with predominance of subtype 1b.
When analyzing the nucleotide sequences of the P-gene region of HBV, no major antiviral drug resistance mutations were detected.
In general, the results of the molecular genetic study conducted in Khabarovsk can significantly supplement the existing ideas about the circulation of HBV and HCV genetic variants on the territory of Russia.
1 World Health Organization. Hepatitis B, Fact Sheet. URL: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (data of access: 07.05.2024); World Health Organization. Hepatitis C, Fact Sheet. URL: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (data of access: 07.05.2024).
2 World Health Organization. Sixty-ninth world health assembly provisional agenda item 15.1. Draft global health sector strategies. Viral hepatitis, 2016–2021.
URL: http://apps.who.int/gb/ebwha/pdf_files/WHA69/A69_32-en.pdf?ua=1 (data of access: 07.05.2024).
3 State report "On the state of sanitary and epidemiological welfare of the population in the Russian Federation in 2022". Мoscow; 2023. URL: https://clck.ru/3FcJ5X
4 Report of the Office of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare in the Khabarovsk Territory "On the state of sanitary and epidemiological well-being of the population in 2022". URL: https://clck.ru/3FcHrj
5 International Committee on Taxonomy of Viruses (ICTV), 2022
URL: https://talk.ictvonline.org/ictv_wikis/flaviviridae/w/sg_flavi/56/hcv-classification (data of access: 15.05.2024).
6 URL: https://www.ncbi.nlm.nih.gov/genbank
7 URL: http://hbv.geno2pheno.org/index.php
8 Report of the Office of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare in the Khabarovsk Territory "On the state of sanitary and epidemiological well-being of the population in 2022". URL: https://clck.ru/3FcHrj
About the authors
Valeriya O. Kotova
Khabarovsk Research Institute of Epidemiology and Microbiology
Author for correspondence.
Email: kotova.valeriya@mail.ru
ORCID iD: 0000-0001-9824-7025
senior researcher, Head, Laboratory of epidemiology and prevention of viral hepatitis and AIDS
Russian Federation, KhabarovskElena A. Bazykina
Khabarovsk Research Institute of Epidemiology and Microbiology
Email: alyonaf@yandex.ru
ORCID iD: 0000-0002-5695-6752
junior researcher, Laboratory of epidemiology and prevention of viral hepatitis and AIDS
Russian Federation, KhabarovskLyudmila A. Balakhontseva
Khabarovsk Research Institute of Epidemiology and Microbiology
Email: adm@hniiem.ru
ORCID iD: 0000-0002-3209-7623
Head, Far Eastern District center for the prevention and control of AIDS
Russian Federation, KhabarovskOlga E. Trotsenko
Khabarovsk Research Institute of Epidemiology and Microbiology
Email: trotsenko_oe@hniiem.ru
ORCID iD: 0000-0003-3050-4472
D. Sci. (Med.), Director
Russian Federation, KhabarovskAnna V. Kuznetsova
Center for Prevention and Combat Against AIDS and Infectious Diseases
Email: cpbsiz.khv@mail.ru
ORCID iD: 0000-0003-3729-4092
Cand. Sci. (Med.), Сhief doctor
Russian Federation, KhabarovskReferences
- Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J. Gastroenterol. 2014;20(18): 5427–34. DOI: https://doi.org/10.3748/wjg.v20.i18.5427
- Kmet Lunacek N., Poljak M., Meglic-Volkar J., et al. Epidemiological, virological and clinical characteristics of hepatitis B virus genotypes in chronically infected persons in Slovenia. Hepat. Mon. 2017;17(3):e43838. DOI: https://doi.org/10.5812/hepatmon.43838
- Martinez M.A., Franco S. Therapy implications of hepatitis C virus genetic diversity. Viruses. 2020;13(1):41. DOI: https://doi.org/10.3390/v13010041
- Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57(3-4):141–50. DOI: https://doi.org/10.1159/000360947
- Lin C.L., Kao J.H. Hepatitis B virus genotypes and variants. Cold Spring Harb. Perspect. Med. 2015;5(5):a021436. DOI: https://doi.org/10.1101/cshperspect.a021436
- Kramvis A., Kew M., François G. Hepatitis B virus genotypes. Vaccine. 2005;23(19):2409–23. DOI: https://doi.org/10.1016/j.vaccine.2004.10.045
- Kramvis A., Kew M.C. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J. Viral. Hepat. 2005;12(5):456–64. DOI: https://doi.org/10.1111/j.1365-2893.2005.00624.x
- Borgia S.M., Hedskog C., Parhy B., et al. Identification of a novel hepatitis C virus genotype from Punjab, India: expanding classification of hepatitis C virus into 8 genotypes. J. Infect Dis. 2018;218(11):1722–9. DOI: https://doi.org/10.1093/infdis/jiy401
- Герасимова В.В., Левакова И.А., Бичурина М.А., Максимова Н.Р. Молекулярно-эпидемиологические особенности вирусного гепатита В. Инфекция и иммунитет. 2015;5(4):297–302. Gerasimova V.V., Levakova I.A., Bichurina M.A., Maksimova N.R. Molecular-epidemiological features of hepatitis B virus. Russian Journal of Infection and Immunity. 2015;5(4):297–302. EDN: https://elibrary.ru/vizekb
- Кашникова А.Д., Быстрова Т.Н., Полянина А.В., Залесских А.А. Молекулярно-генетический мониторинг как компонент эпидемиологического надзора за гепатитом С. Здоровье населения и среда обитания — ЗНиСО. 2022;30(11):76–81. Kashnikova A.D., Bystrova T.N., Polyanina A.V., Zalesskikh A.A. Genetic monitoring as a component of hepatitis C surveillance. Public Health and Life Environment — PH&LE. 2022;30(11):76–81. DOI: https://doi.org/10.35627/2219-5238/2022-30-11-76-81 EDN: https://elibrary.ru/kmaums
- Хорькова Е.В., Лялина Л.В., Микаилова О.М. и др. Актуальные вопросы эпидемиологического надзора за хроническими вирусными гепатитами B, C, D и гепатоцеллюлярной карциномой на региональном уровне. Здоровье населения и среда обитания — ЗНиСО. 2021;29(8):76–84. Khorkova E.V., Lyalina L.V., Mikailova O.M., et al. Current issues of epidemiological surveillance of chronic viral hepatitis B, C, D and hepatocellular carcinoma at the regional level. Public Health and Life Environment — PH&LE. 2021;29(8):76–84. DOI: https://doi.org/10.35627/2219-5238/2021-29-8-76-84 EDN: https://elibrary.ru/mhcuqj
- Соболева Н.В., Карлсен А.А., Кожанова Т.В. и др. Распространенность вируса гепатита С среди условно здорового населения Российской Федерации. Журнал инфектологии. 2017;9(2):56–64. Soboleva N.V., Karlsen A.A., Kozhanova T.V., et al. The prevalence of the hepatitis c virus among the conditionally healthy population of the Russian Federation. Journal Infectology. 2017;9(2):56–64. DOI: https://doi.org/10.22625/2072-6732-2017-9-2-56-64 EDN: https://elibrary.ru/yskwxr
- Колпаков С.Л. Методология изучения эпидемического процесса статистическими и аналитическими методами. Владивосток;2023. Kolpakov S.L. Methodology of Epidemic Process Evaluation Using Statistical and Analytical Methods. Vladivostok;2023. EDN: https://elibrary.ru/fvbtsb
- Кюрегян К.К., Михайлов М.И. Молекулярно-биологические основы контроля вирусных гепатитов. М.;2013. Kyuregyan K.K., Mikhailov M.I. Molecular Biological Basis of Viral Hepatitis Control. Моscow;2013. EDN: https://elibrary.ru/duejiv
- Rajhi M., Ghedira K., Chouikha A., et al. Phylogenetic analysis and epidemic history of hepatitis C virus genotype 2 in Tunisia, North Africa. PLoS One. 2016;11(4):e0153761. DOI: https://doi.org/10.1371/journal.pone.0153761
- Кичатова В.С., Карлсен А.А., Исаева О.В. и др. Лекарственно-резистентные варианты ВГС субтипа 1в, циркулирующие на территории Российской Федерации: анализ аминокислотных мутаций в белках NS5A и Core. Журнал инфектологии. 2018;10(4):30–6. Kichatova V.S., Karlsen A.A., Isaeva O.V., et al. Drug resistant variants of hepatitis c virus genotype 1B in Russia: analysis of amino acid substitutions in NS5A and core proteins. Journal Infectology. 2018;10(4):30–6. DOI: https://doi.org/10.22625/2072-6732-2018-10-4-30-36 EDN: https://elibrary.ru/vvmeki
- Tamura K., Stecher G., Peterson D., et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013; 30(12):2725–9. DOI: https://doi.org/10.1093/molbev/mst197
- Котова В.О., Балахонцева Л.А., Базыкина Е.А. и др. Молекулярно-генетическая характеристика хронического вирусного гепатита В среди пациентов Нанайского района Хабаровского края. Дальневосточный журнал инфекционной патологии. 2018;(35):15–21. Kotova V.O., Bakakhontseva L.A., Bazykina E.A., et al. Molecular-genetic characteristics of chronic viral hepatitis B in patients of the Nanaysky district of the Khabarovsk region. Far Eastern Journal of Infectious Pathology. 2018;(35):15–21. EDN: https://elibrary.ru/yqxrwp
- Котова В.О., Балахонцева Л.А., Базыкина Е.А. и др. Генетическое разнообразие вируса гепатита С среди населения Нанайского района Хабаровского края. Инфекция и иммунитет. 2021;11(1):148–56. Kotova V.O., Balakhontseva L.A., Bazykina E.A., et al. Genetic diversity of hepatitis c virus in Nanaian region, Khabarovsk territory. Russian Journal of Infection and Immunity. 2021;11(1):148–56. DOI: https://doi.org/10.15789/2220-7619-GDO-1265 EDN: https://elibrary.ru/enfery
- Чуланов В.П., Неверов А.Д., Карандашова И.В. и др. Молекулярно-генетические исследования в эпидемиологии вирусных гепатитов: достижения и перспективы. Эпидемиология и инфекционные болезни. Актуальные вопросы. 2014;(2):28–34. Chulanov V.P., Neverov A.D., Karandashova I.V., et al. Molecular genetic studies in the epidemiology of viral hepatitis: progress and prospects. Epidemiology and Infectious Diseases. Current Items. 2014;(2):28–34. EDN: https://elibrary.ru/rzaabv
Supplementary files