Genomic surveillance strategy. Problems and perspectives
- Authors: Akimkin V.G.1, Semenenko T.A.2, Khafizov K.F.1, Ugleva S.V.1, Dubodelov D.V.1, Kolosovskaya E.N.3
-
Affiliations:
- Central Research Institute for Epidemiology
- N.F. Gamaleya National Research Center of Epidemiology and Microbiology
- I.I. Mechnikov North-Western State Medical University
- Issue: Vol 101, No 2 (2024)
- Pages: 163-172
- Section: TOPICAL ISSUES IN SCIENCE
- URL: https://microbiol.crie.ru/jour/article/view/18577
- DOI: https://doi.org/10.36233/0372-9311-507
- EDN: https://elibrary.ru/mymnik
- ID: 18577
Cite item
Abstract
The topic of epidemiologic surveillance is one of the basic concepts in the theory and practice of epidemiologic science. In Russia, generalization of the accumulated factual material and theoretical developments have allowed us to formulate a number of provisions on the nature of the epidemic process. The pandemic of a new coronavirus infection has forced adjustments in all spheres of society, including the activities of the infectious disease epidemiological surveillance system, requiring the development and implementation of innovative solutions. Based on the experience of prompt response to the tasks set by the COVID-19 pandemic, the authors raised the problem of development and implementation of a system of molecular genetic monitoring for pathogens of emerging and re-emerging infections as a priority vector of epidemiological surveillance development.
The introduction of modern molecular biological technologies for the identification of pathogens with epidemic potential, taking into account their genetic diversity, into the system of epidemiologic surveillance has been substantiated based on the experience of using platform solutions created by the Central Research Institute of Epidemiology of Rospotrebnadzor. The strategy of genomic epidemiologic surveillance as a powerful tool to ensure readiness for response measures and management of the epidemic process by implementing and adjusting preventive and anti-epidemic measures was developed.
The Russian platform for aggregation of information on virus genomes (VGARus) developed at the Central Research Institute of Epidemiology of Rospotrebnadzor as a technological, scientific, organizational and infrastructural base of genomic epidemiological surveillance, acting as an interdepartmental consortium, has been introduced into practice. The efficiency of VGARus was shown to assess the mutational variability of SARS-CoV-2, the influence of evolutionary development of circulating pathogens on the characteristics of the epidemic process, the implementation of operational and retrospective analysis of morbidity and prediction of the spread of genetic variants of pathogens.
Full Text
Introduction
The growing threat of epidemic and epizootic outbreaks of new and re-emerging infections, most of which are characterized by sudden onset, high mortality, lack of specific methods of diagnosis and treatment, require the development of new approaches to the organization of epidemiological surveillance. In the context of public health, systemic control and management of the epidemic process of infectious and parasitic diseases are closely related to the problem of biological safety, which is extremely relevant for all countries due to the expanding range of real and potential threats caused by the impact of dangerous biological agents [1–3].
The concept of epidemiological surveillance
The topic of epidemiologic surveillance is one of the basic concepts in the theory and practice of epidemiologic science. In Russia, generalization of the accumulated factual material and theoretical developments allowed us to formulate a number of concepts and provisions on the nature of the epidemic process: the theory of the mechanism of transmission of infectious pathogens by L.V. Gromashevsky [4] and the theory of natural foci of infectious diseases by E.N. Pavlovsky [5]. However, based on these theories, the leading scientists of the country developed the main provisions of epidemiologic surveillance, which had certain differences in interpretation. In the socio-ecological concept of the epidemic process formulated by academician B.L. Cherkassky, epidemiological surveillance was defined as a system of dynamic and comprehensive monitoring for the epidemic process of a particular disease in a certain territory in order to rationalize and improve the effectiveness of preventive and anti-epidemic measures [6]. According to the definition of academician V.I. Pokrovsky, epidemiologic surveillance is the informational support of the infectious disease prevention system, guaranteeing comprehensive tracking of the epidemic process and its determinants (screening) and clearly responding to all possible changes in its development (monitoring) [7].
According to the theory of academician V.D. Belyakov, the basis for the development of the epidemic process are phase changes in the heterogeneity of biological properties of interacting populations of the pathogen and humans, based on feedbacks in the process of self-regulation, with social and natural factors being an important part of it [8]. According to this theory, epidemiologic surveillance is considered as a management system that involves dynamic assessment of trends in the development of the epidemic process in space and time, providing timely intervention in its course in order to reduce the incidence of disease in the general population and eliminate individual infections [9]. In this definition, the goals of epidemiologic surveillance coincide with the goals of the anti-epidemic system as a whole. It is the concept of the self-regulation mechanism of parasitic systems that formed the fundamental basis for the practice of epidemiological surveillance developed in the modern period, which is defined as continuous assessment of the state and trends in the development of the epidemic process for the timely adoption of management decisions that ensure the implementation of measures adequate to the situation.
There are no significant differences in the understanding of the essence of epidemiological surveillance between Russian and foreign experts, but the emphasis is different. Thus, according to the definition of the World Health Organization (WHO), epidemiological surveillance is a systematic continuous collection, comparison, analysis of data and timely dissemination of information among interested parties to make certain decisions [10]. In the domestic health care system, the fact that the object of epidemiological surveillance is the epidemic process, which is a unity of biological, natural and social factors, and the surveillance itself began to be considered as a tool for its cognition, was generally recognized.
Epidemiological surveillance system
The system of epidemiologic surveillance includes three interrelated subsystems: informational, diagnostic and managerial.
The information subsystem is the basic section of epidemiologic surveillance, which takes into account and records data on the status and trends of the epidemic process, causes (biological factor) and conditions (social and natural factors) that support it in a particular territory. Depending on the epidemiological features of the infectious disease, the level of theoretical knowledge and practical capabilities, epidemiological and socio-ecological monitoring are realized.
Diagnostic subsystem provides pre-epidemic diagnostics, including timely detection of preconditions and precursors of epidemic disadvantage, as well as forecasting of further development of the epidemic situation based on the assessment of all information flows.
The management subsystem is focused on the inclusion of information, diagnostic and analytical data in epidemiologic surveillance, taking into account modern achievements of science and practice. Managerial decisions imply drawing up problem-thematic and program-targeted plans, control over their implementation and making adjustments to the tactics of the conducted activities taking into account changing risk factors [11].
Russian platform for aggregation of information on virus genomes
The pandemic of a new coronavirus infection has made adjustments in all spheres of society, including the activities of the health care system. The XII Congress of the All-Russian Public Organization under the name of "All-Russian Scientific and Practical Society of Epidemiologists, Microbiologists and Parasitologists" outlined the main milestones of further development of the sanitary and epidemiological service of the country taking into account the transformation of the general paradigm of epidemiological surveillance and control. Changes in the socio-economic and epidemiological situation in Russia, increasing pressure of adverse environmental factors on humans, urbanization processes, development of new technologies in medicine, food industry and agriculture are reflected in the state of sanitary and epidemiological well-being of the population. Obviously, these trends require fundamentally new approaches to the organization and conduct of epidemiological surveillance of infectious diseases, development of regulatory and legal support, implementation of scientific solutions in practice [12].
The COVID-19 pandemic has vividly demonstrated the devastating consequences of mass infectious diseases that kill millions of people and undermine the global economy. The catastrophic expansion of the incidence of a new coronavirus infection has not only exposed the public health problems of most countries, but has also been an impetus for scientific progress in many fields of medical and biological sciences. The global spread of the new infection caused by SARS-CoV-2 has facilitated the development of innovative tools and technologies in various fields and accelerated the integration of genomic research into public health practice. There is an urgent need to develop new approaches to the organization of epidemiological analysis and forecasting of COVID-19 epidemic development using innovative platform solutions and digital technologies.
The organizational mission to create a system of molecular genetic monitoring during the pandemic COVID-19 belongs to A.Yu. Popova, the head of the Federal Service for Supervision of Consumer Rights Protection and Human Welfare. In accordance with the set tasks and because of the vast experience of anti-epidemic work in Russia, a strategy of proactive response to the spread of a new coronavirus infection was implemented, which made it possible to prevent an excessive burden on the health care system, save millions of lives and prevent large-scale negative consequences for all spheres of life [13, 14].
In accordance with the Resolution of the Government of the Russian Federation dated 23.03. 2021 No. 448 "On Approval of the Temporary Procedure for Provision of Genome Decoding Data of the New Coronavirus Infection Pathogen (COVID-19)" to ensure rapid assessment of the dynamics of the spread of known and new genetic variants of SARS-CoV-2 circulating on the territory of the country, Virus Genome Aggregator of Russia (VGARus), which contains information on nucleotide sequences of SARS-CoV-2 viruses and their mutations circulating in the regions of Russia, was developed and implemented, and can be used for storage, systematization and sampling of data for mutation detection and determination of virus genetic variants. VGARus was developed and consolidated by the Central Research Institute of Epidemiology of Rospotrebnadzor. The software integrated into the VGARus platform allows analyzing sequencing results, determining the probable virus strain, generating standardized reports, and downloading samples for further sequencing [15]. VGARus makes it possible to continuously monitor the mutational variability of SARS-CoV-2, providing essential data for the detection of new genetic variants and tracking their prevalence in Russia. The algorithm of work with VGARus data allows to carry out operative and retrospective analysis of distribution of genetic variants of SARS-CoV-2 taking into account the latest information on genetic diversity of COVID-19 pathogen.
The main purpose of VGARus is to centralize the collection and analysis of the dynamics and structure of identified SARS-CoV-2 variants in Russia [16]. Currently, all Russian scientific institutions involved in coronavirus genome sequencing and registered as users on the portal have the opportunity to upload the studied genomic sequences to VGARus. The obtained registration certificates allow its participants to use information from the national database. The platform is accessed through the portal genome.crie.ru.
Thus, a scientific consortium was, in fact, created, which included institutions of Rospotrebnadzor, the Ministry of Health of the Russian Federation and a number of other agencies. Currently, more than 150 organizations are integrated into the VGARus system; a significant part of them perform mass sequencing of SARS-CoV-2 genomes and download sequences. Preliminary quality assessment of these samples is performed, which typically includes analysis using polymerase chain reaction to determine viral load and assess the suitability of the sample for next-generation whole-genome sequencing. Algorithms running in the database automatically perform mutation analysis and identification of the SARS-CoV-2 variant in each sample. After loading the nucleotide sequence of the virus, the system automatically starts the process of sequencing validation, analyzing the belonging to one or another genetic variant. The downloaded genomic information is processed using Pangolin and V-TRACE programs (developed by specialists of the Central Research Institute of Epidemiology of Rospotrebnadzor) in automated mode with subsequent bioinformatic analysis.
Each sample in the system contains not only the nucleotide sequence and technical data, but also information on the place and time of collection of biological material, as well as data on the examined person: sex, region, age, vaccination status, estimated number of contacts, comorbidities, recent foreign travel, etc. These data, given their epidemiologic significance, will allow them to be used for operational and retrospective analysis. When samples are registered in the database, they will automatically receive an internal registration number, after which the SARS-CoV-2 genome sequence can be added to the sample information field. Technical information also includes data on the organizations involved in the collection, laboratory sample preparation, the date the sample was sent and received, the date the information was entered, and the date the current record was generated in the system [17–20].
Molecular genetic monitoring during the COVID-19 pandemic
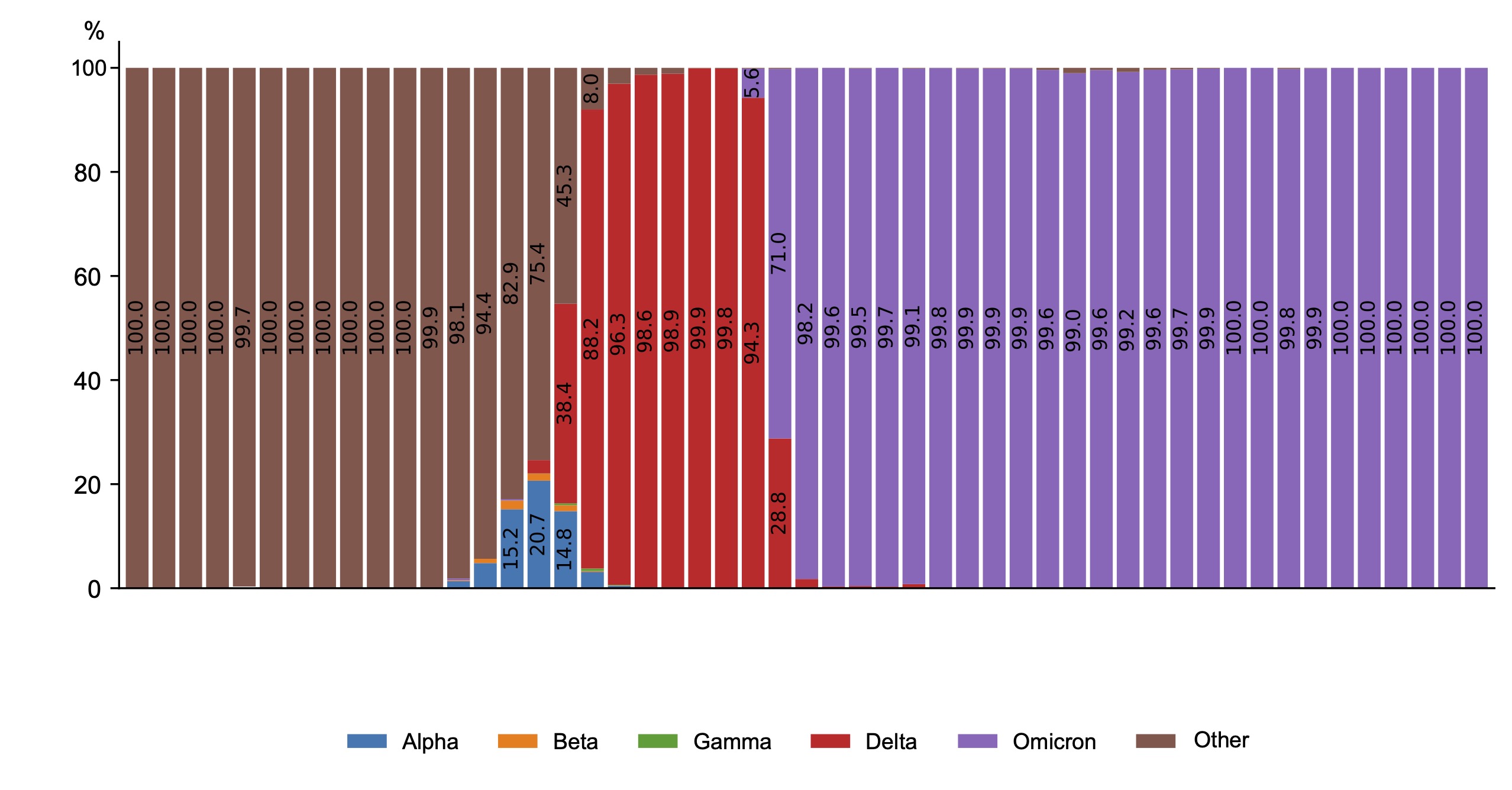
The national VGARus database, which contains a large set of SARS-CoV-2 sequences, is an invaluable resource for tracking and deciphering the evolution of the COVID-19 pandemic (Fig. 1).
Fig. 1. Dynamics of SARS-CoV-2 genetic variants in Russia in 2020–2024.
During the first year of SARS-CoV-2 presence in the human population, no nucleotide substitutions in its genome that could significantly alter the pathogen's properties were detected. However, since preservation of the pathogen as a biological species is impossible without evolutionary development, the heterogeneity of the coronavirus population began to increase due to circulation of different variants with subsequent selection and formation of the epidemic variant of the pathogen. Dynamic monitoring of mutational variability of circulating coronaviruses started in Russia in December 2020. During this period, a genetic variant initially known as British was discovered, which was later renamed Alpha (B.1.1.7) in accordance with the WHO decision to abandon the use of country names to designate strains. Among the mutations found in the S-protein gene, the most significant were N501Y, P681H, and Δ69-70, which affect the transmissibility of the virus and its ability to infect cells and evade the immune response. The discovery of the Alpha variant in Russia coincided with an increase in the number of cases in late 2020 and early 2021. Later, the Beta variant (B.1.351), first detected in the Republic of South Africa, and Gamma (P.1), identified in Brazil, were identified, but they were not widespread in Russia, accounting for only a small percentage of the total number of new cases. The Alpha variant spread across Russia in winter 2021, and in summer 2021, a new variant, Delta (B.1.617.2), emerged, which was accompanied by a significant increase in the number of cases and hospitalizations, a severe course of coronavirus infection and high mortality rates. The Delta genetic variant prevailed in Russia from May to December 2021, with its share among the identified variants approaching 100%. The dominant variant in all months of observation since the start of registration of genetic variant Delta was the variant, which was named AY.122 by the Pangolin classifier from 26.11.2021 (83.3%). In total, more than 400 sublineages of the Delta genetic variant were isolated in Russia.
The process of change of biological properties of SARS-CoV-2 virus with the subsequent change of prevailing genetic variants is associated with the evolution of the virus and the formation of its epidemic variant with a natural change in the immunological structure of the human population in the chain of circulation of the pathogen. As a result of changes in the population of SARS-CoV-2 virus during circulation of the Delta genetic variant (increased virulence, increased abundance), a new rise in morbidity among the population was observed. Going by the theory of self-regulation of parasitic systems, the process went from the phase of reservation to the phase of epidemic transformation and then epidemic spread. Changes in the genetic properties of the virus led to changes in its pathogenicity and, as a result, influenced the severity of the clinical course of the disease and the characteristics (indicators) of the manifestations of the epidemic process.
As a result of the interaction between the pathogen and human populations, taking into account their heterogeneity, a new variant of SARS-CoV-2 coronavirus, first identified in Botswana and South Africa, was identified in late 2021 and named Omicron (B.1.1.529 according to the PANGO classification) by WHO. The Omicron variant started to spread rapidly from December 2021 and is now completely dominant in Russia (100% of samples tested). Analysis of VGARus data revealed dissociation of the Omicron genetic lineage in Russia with the highest frequency of circulation of subvariants BA.1 (54.5%), BA.1.1.1 (21.6%) and BA.2 (23.8%).
Spring 2022 was a period of epidemiological well-being characterized by a low incidence of COVID-19. However, the emergence of Omicron subvariants BA.4 and especially BA.5 led to an increase in incidence that lasted until the end of October 2022. In late 2022 and early 2023, highly transmissible variants such as BQ.1* (subvariant BA.5) emerged, indicating the dynamic and complex nature of SARS-CoV-2 evolution. The early months of 2023 saw a resurgence of new forms of old strains, including Omicron BA.2, which returned as recombinant forms of XBB*, being prevalent for most of 2023. Within the XBB lineage, its own prevalent forms emerged, such as XBB.1.5 (Kraken), XBB.1.16 (Arcturus), and XBB.1.9.2.1 (EG.5, Eris). The emergence of the latter coincided with the beginning of the increase in the incidence of the disease in the country in September 2023.
At the end of August 2022, a new SARS-CoV-2 variant, BA.2.86 (Pirola), which had many additional mutations compared to previous Omicron variants, was first detected in Denmark and Israel. However, it was not the cause of the increase in incidence in many countries, because the growth itself began earlier than the active spread of this genetic variant. The first cases of BA.2.86 infection were first detected in Russia in early November 2023, and further spread of BA.2.86 could slightly prolong the stage of the rise, including in January-February 2024, when the BA.2.86 JN.1 subvariant began to spread actively, becoming prevalent both in Russia and practically in all other countries. In total, more than 600 different Omicron sublineages have been registered in Russia, although this number may vary depending on the definition of the sublineage (Fig. 2).
Fig. 2. Dynamics of Omicron genetic variant in Russia in 2022–2024.
SARS-CoV-2 has evolved and genomic changes have led to the emergence of such characteristics as more intense transmission, changes in clinical symptoms, evasion of the immune response, and drug resistance. The phase self-reorganization of the pathogen population during the emergence of the Omicron genetic variant led to a decrease in its virulence, which was accompanied by a decrease in the severity of diseases, the number of hospitalized and deceased patients, which may indicate a phase of reserve transformation in accordance with the theory of self-regulation of parasitic systems. Preservation of the pathogen as a biological species is impossible without evolutionary development, which is facilitated by genome instability and mutations, as well as expansion of the range of heterogeneity of the SARS-CoV-2 virus population due to circulation of both low- and high-virulence variants with subsequent stabilizing selection and formation of the epidemic variant of the pathogen. Therefore, the phase of reservation always balances on the border with the phase of epidemic transformation, when new strains capable of bypassing the protection previously formed by the human population, eluding protection from vaccines and post-infection immunity, emerge and gain advantage in natural selection.
The importance of genomic epidemiologic surveillance
Molecular genetic monitoring makes it possible to anticipate changes in phenotypic properties affecting the epidemic process manifestation indicators and socio-economic consequences of such changes based on a more detailed study of the genetic characteristics of pathogens.
It is important to note that at the end of 2023, the volume of genomic sequencing has decreased worldwide, and this could possibly lead to the emergence of new variants without their detection for an extended period of time. A significant amount of data has been accumulated on evolutionary changes in the SARS-CoV-2 genome, taking into account new epidemiologic properties. Monitoring and genomic sequencing of the virus are important for the identification of new genetic variants and the development of public health strategies.
Genomic sequencing is increasingly being used to collect data on other pathogens, to investigate outbreaks of severe infectious diseases (cholera, Ebola, Dengue, bacterial meningitis, poliomyelitis, etc.) that may lead to public health emergencies, and to provide emergency medical care for critical epidemiologic situations. For example, the Global Influenza Surveillance and Response System (GISRS) routinely uses genomic sequencing as an integral part of the zoonotic influenza outbreak response and pandemic preparedness package, and for seasonal influenza surveillance to develop seasonal vaccine recommendations and monitor antiviral susceptibility. GISRS has been used to incorporate SARS-CoV-2 virus into sentinel surveillance systems for influenza-like illnesses, acute respiratory infections, and severe acute respiratory infections to collect data to inform countries' national COVID-19 pandemic policies and responses1.
At the 74th Session of the World Health Assembly held on 30.05.2021, WHO Member States called for strengthening the role of genomic epidemiological surveillance in emergency preparedness and response, but there are problems related to the lack of necessary capacity and appropriate infrastructure for laboratory research. Only two-thirds of countries, including Russia, have the capacity to conduct genomic sequencing and ensure regular use of this powerful technology, while the remaining countries are only establishing genomic surveillance systems for pathogens with epidemic potential [21].
The 2022-2032 Surveillance Strategy, developed by WHO based on previous experience and lessons learned from the COVID-19 pandemic, relies on the special role of genomics in public health, which it plays due to the ability to utilize the results of genomic research in various fields of medicine. The strategy is not limited to a single pathogen or a specific epidemiologic threat. Rather, it aims to develop a unified vision for using genomics as a powerful complementary tool to address the public health challenges of preparedness and response to pandemics and epidemics across a broad spectrum.
Conclusion
The new Global Strategy for Genomic Epidemiological Surveillance of Pathogens with Pandemic and Epidemic Potential notes that genomic epidemiological surveillance is making a significant difference to public health by providing a better understanding of the nature, evolution and pathways of infectious pathogens. Genomic data on pathogens with pandemic and epidemic potential, combined with clinical, epidemiologic and other data, are being used for risk assessment, development of vaccines, drugs and diagnostic tests, and decision-making on epidemiologic and social control measures. New technologies in sequencing and bioinformatics that have emerged in recent years have enabled a number of countries to make significant progress in building and strengthening their capabilities in this area.
The goal of genomic epidemiologic surveillance is to manage the epidemic process on the basis of systemic data on changes in the genetic properties of infectious pathogens with significant epidemic (pandemic) potential.
Objectives of genomic epidemiologic surveillance:
- Operational and retrospective analysis of changes in the genetic properties of circulating and emerging pathogen variants, allowing dynamic tracking of the change of dominant gene variants.
- Assessment of the influence of the structure of circulating pathogens taking into account the peculiarities of territorial distribution on the characteristics of the epidemic process.
- Identification of predictors of the unfavorable epidemiological situation development on the basis of molecular genetic monitoring.
- Predicting the development of the epidemic process of infectious diseases based on knowledge of changes in the genetic properties of the pathogen using innovative platform solutions and application of digital technologies.
- Searching for and predicting new human and animal pathogens.
- Developing rapid responses to emerging infections of epidemic and pandemic potential.
- Management of the epidemic process through the development of implementation and adjustment of the system of preventive and anti-epidemic measures.
- Creation of innovative vaccine and drug products.
Taking into account the above mentioned, Russia has made a significant step forward in the development of this scientific field. Genomic epidemiologic surveillance is a qualitatively new level of epidemiologic surveillance, taking into account the possibilities of studying genetic properties of infectious pathogens.
Thus, genomic epidemiologic surveillance, based on the knowledge of molecular genetic properties of infectious pathogens, is an essential component of biosecurity of the Russian Federation and a strategic area of scientific and technological development2.
1 WHO. Global Influenza Surveillance and Response System (GISRS). URL: https://who.int/initiatives/global-influenza-surveillance-and-response-system (date of access: 31.01.2024).
2 Decree of the President of the Russian Federation of 28.02.2024 No. 145 "On the Strategy of Scientific and Technological Development of the Russian Federation".
About the authors
Vasily G. Akimkin
Central Research Institute for Epidemiology
Email: uglevas@bk.ru
ORCID iD: 0000-0003-4228-9044
D. Sci. (Med.), Prof., Full Member of RAS, Director
Russian Federation, MoscowTatyana A. Semenenko
N.F. Gamaleya National Research Center of Epidemiology and Microbiology
Email: uglevas@bk.ru
ORCID iD: 0000-0002-6686-9011
D. Sci. (Med.), Professor, Head, Department of epidemiology
Russian Federation, MoscowKamil F. Khafizov
Central Research Institute for Epidemiology
Email: uglevas@bk.ru
ORCID iD: 0000-0001-5524-0296
Cand. Sci. (Biol.), Head, Genomic research laboratory
Russian Federation, MoscowSvetlana V. Ugleva
Central Research Institute for Epidemiology
Author for correspondence.
Email: uglevas@bk.ru
ORCID iD: 0000-0002-1322-0155
D. Sci. (Med.), Assoc. Prof., Head, Scientific and analytical department
Russian Federation, MoscowDmitry V. Dubodelov
Central Research Institute for Epidemiology
Email: uglevas@bk.ru
ORCID iD: 0000-0003-3093-5731
Cand. Sci. (Med.), senior researcher, Laboratory of viral hepatitis, Department of molecular diagnostics and epidemiology
Russian Federation, MoscowElena N. Kolosovskaya
I.I. Mechnikov North-Western State Medical University
Email: uglevas@bk.ru
ORCID iD: 0000-0001-6667-2377
D. Sci. (Med.), Professor, Department of epidemiology, parasitology and disinfectology
Russian Federation, St. PetersburgReferences
- Акимкин В.Г., Попова А.Ю., Плоскирева А.А. и др. COVID-19: эволюция пандемии в России. Сообщение I: проявления эпидемического процесса COVID-19. Журнал микробиологии, эпидемиологии и иммунобиологии. 2022;99(3):269–86. Akimkin V.G., Popova A.Yu., Ploskireva A.A., et al. COVID-19: the evolution of the pandemic in Russia. Report I: Manifestations of the COVID-19 epidemic process. Journal of Microbiology, Epidemiology and Immunobiology. 2022;99(3):269–86. doi: https://doi.org/10.36233/0372-9311-276 EDN: https://elibrary.ru/zxgtfd
- Акимкин В.Г., Кузин С.Н., Колосовская Е.Н. и др. Характеристика эпидемиологической ситуации по COVID-19 в Санкт-Петербурге. Журнал микробиологии, эпидемиологии и иммунобиологии. 2021;98(5):497–511. Akimkin V.G., Kuzin S.N., Kolosovskaya E.N., et al. Assessment of the COVID-19 epidemiological situation in St. Petersburg. Journal of Microbiology, Epidemiology and Immunobiology. 2021;98(5):497–511. doi: https://doi.org/10.36233/0372-9311-154 EDN: https://elibrary.ru/dtmnhz
- Семененко Т.А. Роль банка сывороток крови в системе биологической безопасности страны. Вестник Росздравнадзора. 2010;(3):55–8. Semenenko T.A. The role of the blood serum bank in the biological safety system of the country. Bulletin of Roszdravnadzor. 2010;(3):55–8. EDN: https://elibrary.ru/muutej
- Громашевский Л.В. Общая эпидемиология. М.;1965. Gromashevskii L.V. General Epidemiology. Moscow;1965.
- Павловский Е.Н. Природная очаговость трансмиссивных болезней в связи с ландшафтной эпидемиологией зооантропонозов. М.;1964. Pavlovskii E.N. Natural foci of Vector-Borne Diseases in Connection with the Landscape Epidemiology of Zooanthroponoses. Moscow;1964. EDN: https://elibrary.ru/zgmuqp
- Черкасский Б.Л. Теоретическое обоснование структуры эпидемиологического надзора. В кн.: Покровский В.И., ред. Эпидемиологический надзор за инфекционными болезнями. М.;1987. Cherkasskii B.L. Theoretical substantiation of the structure of epidemiological surveillance. In: Pokrovskii V.I., ed. Epidemiological Surveillance of Infectious Diseases. Moscow;1987.
- Покровский В.И. Пути оптимизации эпидемиологического надзора за инфекционными болезнями в стране. Журнал микробиологии, эпидемиологии и иммунобиологии. 1986;63(11):3–7. Pokrovskii V.I. Ways to optimize epidemiological surveillance of infectious diseases in the country. Journal of Microbiology, Epidemiology and Immunobiology. 1986;63(11):3–7. EDN: https://elibrary.ru/pioohv
- Беляков В.Д. Общие закономерности функционирования паразитарных систем (механизмы саморегуляции). Паразитология. 1986;20(4):249–55. Belyakov V.D. General patterns of functioning of parasitic systems (mechanisms of self-regulation). Parasitology. 1986;20(4):249–55.
- Беляков В.Д. Эпидемиологический надзор – основа современной организации противоэпидемической работы. Журнал микробиологии, эпидемиологии и иммунобиологии. 1985;62(5):53–8. Belyakov V.D. Epidemiological surveillance is the basis of modern organization of anti-epidemic work. Journal of Microbiology, Epidemiology and Immunobiology. 1985;62(5):53–8. EDN: https://elibrary.ru/zfxutr
- WHO. Strengthening Population Health Surveillance: A Tool for Selecting Indicators to Signal and Monitor the Wider Effects of the COVID-19 Pandemic. Copenhagen; 2021.
- Фельдблюм И.В. Эпидемиологический надзор за вакцинопрофилактикой. Журнал МедиАль. 2014;(3):37–55. Fel'dblyum I.V. Epidemiologic surveillance over preventive vaccination. Medial Journal. 2014;(3):37–55. EDN: https://elibrary.ru/sxhknx
- Попова А.Ю., Акимкин В.Г., ред. Материалы XII Съезда Всероссийского научно-практического общества эпидемиологов, микробиологов и паразитологов. М.;2022. Popo- va A.Yu., Akimkin V.G., eds. Proceedings of the XII Congress of the All-Russian Scientific and Practical Society of Epidemiologists, Microbiologists and Parasitologists. Moscow;2022. doi: https://doi.org/10.36233/978-5-6048873-1-8 EDN: https://elibrary.ru/nrlneo
- Стародубов В.И., Береговых В.В., Акимкин В.Г. и др. COVID-19 в России: эволюция взглядов на пандемию (часть 1). Вестник Российской академии медицинских наук. 2022;77(3):199–207. Starodubov V.I., Beregovykh V.V., Akimkin V.G., et al. COVID-19 in Russia: evolution of views on the pandemic. Report I. Annals of the Russian Academy of Medical Sciences. 2022;77(3):199–207. DOI: https://doi.org/10.15690/vramn2118 EDN: https://elibrary.ru/sqglyh
- Акимкин В.Г., Попова А.Ю., Хафизов К.Ф. и др. COVID-19: эволюция пандемии в России. Сообщение II: динамика циркуляции геновариантов вируса SARS-CoV-2. Журнал микробиологии, эпидемиологии и иммунобиологии. 2022;99(4):381–96. Akimkin V.G., Popova A.Yu., Khafizov K.F., et al. COVID-19: the evolution of the pandemic in Russia. Report II: dynamics of the circulation of SARS-CoV-2 genetic variants. Journal of Microbiology, Epidemiology and Immunobiology. 2022;99(4):381–96. doi: https://doi.org/10.36233/0372-9311-295 EDN: https://elibrary.ru/kvulas
- Латыпова М.Ф., Цибин А.Н., Комаров А.Г. и др. Организация геномного надзора за SARS-CoV-2 в структуре Департамента здравоохранения города Москвы. Проблемы социальной гигиены, здравоохранения и истории медицины. 2022;30(S):1061–6. Latypova M.F., Tsibin A.N., Komarov A.G., et al. Organization of genomic surveillance for SARS-CoV-2 within the Moscow City Health Department. Problems of Social Hygiene, Public Health and History of Medicine, Russian Journal. 2022;30(S):1061–6. DOI: https://doi.org/10.32687/0869-866X-2022-30-s1-1061-1066 EDN: https://elibrary.ru/mshhnn
- Акимкин В.Г., Семененко Т.А., Углева С.В. и др. COVID-19 в России: эпидемиология и молекулярно-генетический мониторинг. Вестник Российской академии медицинских наук. 2022;77(4):254–60. Akimkin V.G., Semenenko T.A., Ugleva S.V., et al. COVID-19 in Russia: epidemiology and molecular genetic monitoring. Annals of the Russian Academy of Medical Sciences. 2022;77(4):254–60. doi: https://doi.org/10.15690/vramn2121 EDN: https://elibrary.ru/dozijs
- Akimkin V.G., Semenenko T.A., Ugleva S.V., et al. COVID-19 epidemic process and evolution of SARS-CoV-2 genetic variants in the Russian Federation. Microbiol. Res. 2024;15(1):213–24. DOI: https://doi.org/10.3390/microbiolres15010015
- Стародубов В.И., Береговых В.В., Акимкин В.Г. и др. COVID-19 в России: эволюция взглядов на пандемию. Сообщение 2. Вестник Российской академии медицинских наук. 2022;77(4):291–306. Starodubov V.I., Beregovykh V.V., Akimkin V.G., et al. COVID-19 in Russia: evolution of views on the pandemic. Report II. Annals of the Russian Academy of Medical Sciences. 2022;77(4):291–306. DOI: https://doi.org/10.15690/vramn2122 EDN: https://elibrary.ru/ojjdra
- Акимкин В.Г. Эпидемиология и диагностика COVID-19. Мониторинг эволюционных изменений вируса SARS-CoV-2. Вестник Российской академии наук. 2022;92(7): 647–53. Akimkin V.G. COVID-19 epidemiology and diagnosis: monitoring evolutionary changes in the SARS-COV-2 virus. Herald of the Russian Academy of Sciences. 2022;92(7):647–53. DOI: https://doi.org/10.31857/S0869587322070027 EDN: https://elibrary.ru/nrfrlg
- Хафизов К.Ф., Петров В.В., Красовитов К.В. и др. Экспресс-диагностика новой коронавирусной инфекции с помощью реакции петлевой изотермической амплификации. Вопросы вирусологии. 2021;66(1):17–28. Khafizov K.F., Petrov V.V., Krasovitov K.V., et al. Rapid diagnostics of novel coronavirus infection by loop-mediated isothermal amplification. Problems of Virology. 2021;66(1):17–28. doi: https://doi.org/10.36233/0507-4088-42 EDN: https://elibrary.ru/uklaki
- Carter L.L., Yu M.A., Sacks J.A., et al. Global genomic surveillance strategy for pathogens with pandemic and epidemic potential, 2022–2032. Bull. World Health Organ. 2022;100(4):239-A. DOI: https://doi.org/10.2471/blt.22.288220