An integrative approach to assessing the pathogenic potential of Escherichia coli strains isolated from urine
- Authors: Makarova M.A.1,2, Matveeva Z.N.1, Kaftyreva L.A.1,2
-
Affiliations:
- Saint-Peterburg Pasteur Institute
- North-Western State Medical University named after I.I. Mechnikov
- Issue: Vol 101, No 1 (2024)
- Pages: 72-79
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18554
- DOI: https://doi.org/10.36233/0372-9311-493
- EDN: https://elibrary.ru/ytaqsf
- ID: 18554
Cite item
Abstract
Introduction. Uropathogenic Escherichia coli (UPEC) are characterized by the ability to survive and reproduce in the urinary tract due to the presence of specific virulence factors. In routine laboratory practice, the detection of diagnostically significant bacteriuria does not provide an idea of the level of infection of the urinary system (renal parenchyma, bladder), the pathogenic potential of the strain in the progression and chronicity of the infectious process, and the occurrence of life-threatening conditions (urosepsis, meningitis).
Objective. To characterize the population structure, genetic diversity and pathogenic potential of E. coli strains isolated from urine.
Materials and methods. 194 strains of E. coli isolated from urine were studied. Detection of 17 genes encoding the synthesis of: adhesins (pap, fimH, sfa, focG, afa), toxins (hlyA, cvaC, cnf, cdtB), capsular antigens (kpsMTII, kpsMTIII, kpsMT K1), siderophores (fyuA, iutA), invasins (ibeA), genetic markers of the pathogenicity island (PAI) of UPEC CFT073, the gene (traT) encoding serum resistance capacity and phylogenetic groups were performed by PCR (CXT-1000, BioRad, USA) with published primers (Synthol, Sibenzyme, Evrogen, Russia). To assess the statistical significance of differences, Fisher's exact test was used. Differences were considered significant at a confidence interval of 95% (p < 0.05).
Results. E. coli strains more often (p < 0.05) belonged to the phylogenetic group B2 (57.7%). Pathogenetically significant virulence determinants were identified in 97.9% of strains. Based on the combination of 17 genes, 134 individual virulence genotypes were identified. In 93.3% of strains, a genetic predisposition to the occurrence of recurrent urinary tract infections (UTIs) was revealed, in 6.9% there was a potential for the development of pyelonephritis and recurrent cystitis. Markers of life-threatening complications of UTI were identified in 12% of strains, of which 10.7% were the development of urosepsis and 1.3% were meningitis.
Conclusion. Detection of a complex of genes in E. coli strains isolated from urine confirms the etiological significance of the isolate and allows one to assess the pathogenic potential for the development of chronic and severe life-threatening complications.
Full Text
Introduction
The urinary tract is a common locus of bacterial infection, and Escherichia coli is the most frequent pathogen in this biotope. Urinary tract infections (UTIs) can present with a variety of clinical presentations ranging from asymptomatic bacteriuria, ascending infections (acute pyelonephritis) to severe urosepsis [1-3].
Uropathogenic E. coli (UPEC) are characterized by an increased adaptive ability to survive and multiply in the urinary tract due to the presence of specific lipopolysaccharides, capsules, outer membrane proteins, fimbriae, pili, secreted toxins, siderophores, as well as serum resistance. The uropathogenic potential of E. coli is consistently realized at different stages of the infection process: adhesion, colonization and persistence [4].
Specific adhesins are required for successful colonization of the urinary system tissues by E. coli. The major adhesion factors include pili or fimbriae. Type 1 pili (FimH), P, S, and F1C fimbriae are typical for UPEC. FimH adhesins, encoded by the fimH gene, play an important role in the initiation of UTI development and are considered the most common virulence factor of UPEC [5, 6]. Strains causing cystitis always express type 1 fimbriae; in the absence of other fimbriae, infection is limited to bladder lesions. The major UPEC-specific pili are the P-fimbriae encoded by the pap gene. They are absent in commensal and diarrheagenic E. coli and were named this way because they are more frequently found in strains associated with pyelonephritis [7]. Mannose-resistant S pili are categorized into Sfa-, F1C-pili (Foc) and S/F1C-related pili (Sfr). These adhesins have a high degree of homology but differ in receptor specificity; S-pili are expressed predominantly by sepsis- (SEPEC) and neonatal meningitis-causing E. coli (NMEC) strains, but may also occur in UPEC strains causing ascending UTIs. In addition to fimbrial adhesins, afimbrial adhesins encoded by the afa gene, which promote adhesion to urothelial cells, are widely distributed in UPEC. Strains synthesizing afibrial adhesins have a high potential for the development of pyelonephritis and recurrent cystitis [8].
Toxins play an important role in UTIs because they promote bacterial dissemination in tissues, increase cytotoxicity, neutrophil resistance, as well as damage and disrupt host cell metabolism. The most studied toxin secreted by UPEC is α-hemolysin, HlyA (a product of the hlyA gene), which not only stimulates apoptosis of target cells, including neutrophils, T lymphocytes, and renal epitheliocytes, but also causes degradation of regulatory and structural components of the cytoskeleton, promoting bladder cell exfoliation and phagocyte destruction. CNF1 (cytotoxic necrotic factor 1) toxin encoded by the cnf gene inhibits polymorphonuclear phagocytosis, promotes the production of bioactive components, causing functional and structural damage as well as apoptosis of bladder epithelial cells [6, 8]. A toxin with DNAase activity, CDT (cytolethal toxin expansion factor), encoded by the cdt gene, leads to cell apoptosis; it is found in more than 90% of UPEC strains [9, 10].
Factors of UPEC persistence include capsules (synthesis of K-antigens), which protect the bacteria from phagocytosis and the bactericidal effect of the complement system [8].
The production of siderophores (iron-binding proteins), which determine the ability of bacterial cells to capture iron, is crucial for the survival of UPEC in the urethral tract. Synthesis of siderophores increases the virulence of UPEC. The main siderophores include aerobactin (iutA) and yersinebactin (fyuA) [11].
In the routine practice of bacteriologic examination, methods for typing E. coli causing UTIs are not used. The criterion for the diagnosis of UTI is the detection of microorganisms at a concentration of at least 103 CFU in 1 ml of urine. The detection of diagnostically significant bacteriuria does not give an idea of the localization of infection of the urinary system (renal parenchyma, bladder) [1]. Therefore, one of the most important tasks of the clinical microbiology laboratory is a meaningful analysis of the results obtained, as well as an assessment of the etiologic significance of the isolated microorganism [12]. The main difficulty in interpreting the results is represented by isolates from urine specimens, due to the possible contamination of E. coli by representatives of intestinal microflora. The microbiologist must not only determine whether the E. coli isolate is the true pathogen or a consequence of contamination of the sample at the pre-analytical stage, but also assess the pathogenic potential of a particular isolate in the development of chronic UTIs or severe life-threatening conditions such as sepsis or meningitis. Although research has been ongoing for many years, specific criteria for categorizing strains as UPEC have not been established.
As a result of the above, the aim of this work is to characterize the population structure, genetic diversity and pathogenic potential of E. coli strains isolated from urine.
Materials and methods
The objects of the study were 194 E. coli strains isolated from the urine of patients with uncomplicated UTIs. The subject of the study was biological properties of the strains reflecting pathogenicity (virulence genes associated with adhesion, invasion, toxin formation, persistence, etc.).
The study was conducted with the informed consent of the patients. The research protocol was approved by the Ethics Committee of the Saint-Peterburg Pasteur Institute (protocol No. 27, July 2, 2020).
DNA isolation was performed using the InstaGeneTMMatrix kit (BioRad). PCR was performed in automatic amplification mode in the thermocycler СХТ-1000 (BioRad). We used a ready mix with Taq DNA polymerase, PCR Master Mix (ThermoFisher Scientific). The primers were added between 0.5–1.5 μl with unchanged sample volume (20 μl), which was achieved by changing the volume of sterile distilled water accordingly. Previously studied primers (Syntol, Sibenzyme, Eurogen) were used [13–15]. The amplified DNA fragments were separated in 0.5×TBE buffer with ethidium bromide at 120 V for 60 min in a horizontal gel electrophoresis chamber (BioRad). GelDoc documentation system (BioRad) was used to visualize PCR results. DNA marker 100 bp + 1.5 Kb + 3 Kb (Sibenzyme) was used as molecular weight markers.
Strains were tested for the presence of 17 genes encoding the synthesis of virulence factors: Adhesins (pap, fimH, sfa, focG and afa), toxins (hlyA, cvaC, cnf and cdtB), capsule antigens (kpsMTII, kpsMTIII, kpsMT K1), siderophores (fyuA and iutA), invasins (ibeA), as well as for the presence of genetic markers of pathogenicity island (PAI) UPEC CFT073 and gene (traT) encoding a serum resistance.
Phylogenetic classification of E. coli strains was performed by multiplex phylotyping based on PCR using primers targeting three markers: chuA, yjaA, and TspE4.C2 [16].
The obtained data were processed using the Excel computer program (Microsoft). Fisher’s exact test was used to assess the statistical significance of differences in mean values. Differences were considered statistically significant when the confidence interval was 95% (p < 0.05).
Results
When analyzing the combination of chuA, yjaA, and TspE4.C2 genes, it was found, statistically, that E. coli strains were significantly more frequent (p < 0.05) in phylogenetic group B2 (57.7%) compared to strains of phylogenetic groups A (4.6%), B1 (7.2%), and D (30.4%); significantly less frequent (p < 0.05) in phylogenetic groups A and B1 compared to B2 and D.
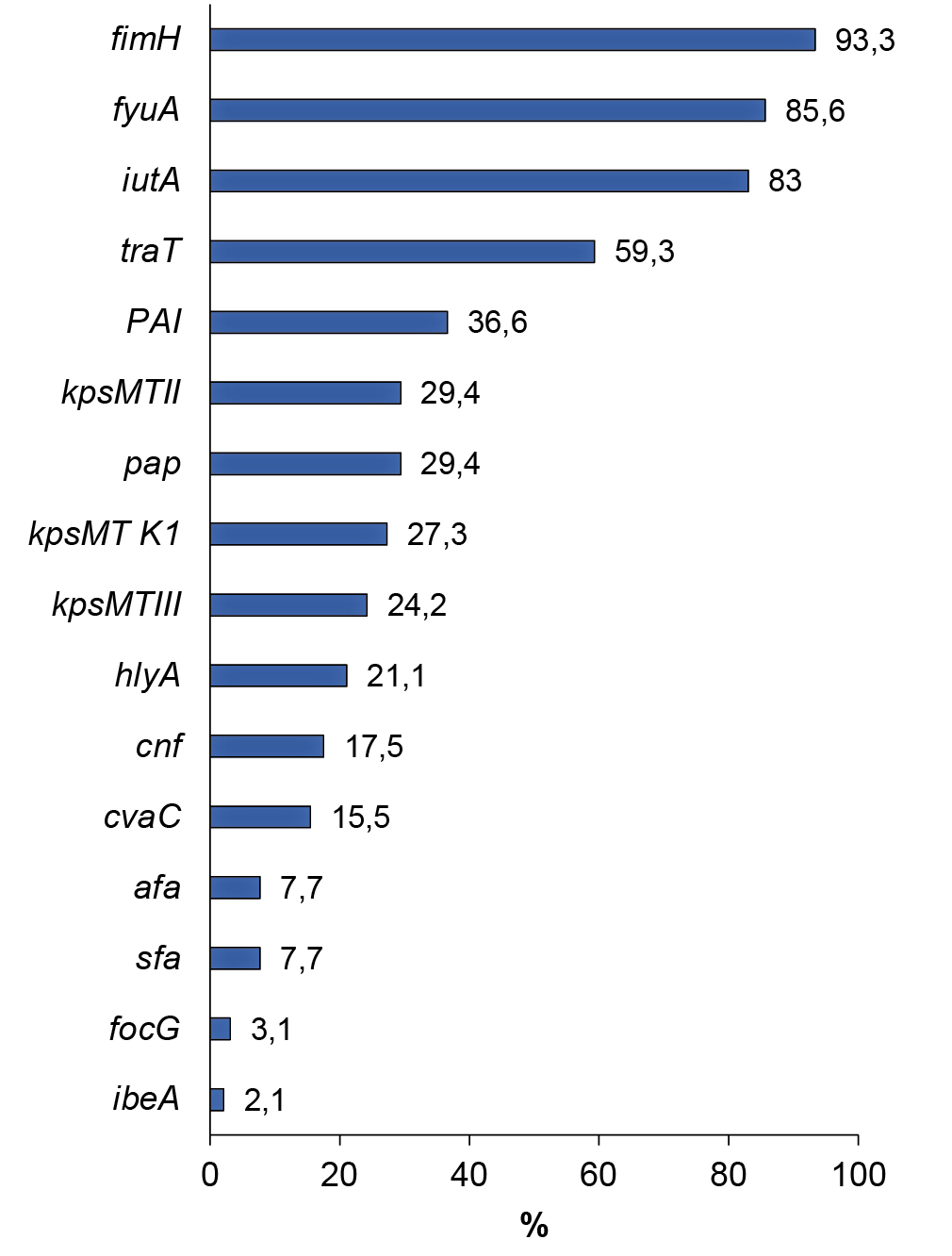
The prevalence of genes encoding the synthesis of UPEC virulence factors ranged from 2.1% (ibeA) to 93.3% (fimH). The gene responsible for the production of cytolethal expanding toxin (cdtB), involved in the suppression of cell proliferation with subsequent cell death, was not identified in any of the strains. The frequency of occurrence of genetic determinants encoding UPEC virulence factors is shown in the figure below.
Prevalence of genes encoding UPEC virulence factors.
Analysis of the detection of genes associated with adhesion showed that almost all strains contained the fimH gene (93.3%) encoding mannose-sensitive type I fimbriae. The pap gene, responsible for the synthesis of pyelonephritis-associated pili, was found in almost every third strain (29.4%); the afa gene, encoding afimbrial adhesins, was found in 7.7% of the studied E. coli strains; sfa and focG determinants associated with fimbrial adhesins — in 7.7 and 3.1%; hlyA and cnf genes encoding synthesis of toxins (α-hemolysin and cytonecrotic factor) — in 21.1 and 17.5%; cvaC gene responsible for colicin V production — in 15.5%. The ibeA gene responsible for invasion of endothelial cells of the blood-brain barrier was found in 4 (2.1%) E. coli strains. The frequency of occurrence of genetic determinants encoding synthesis of siderophores: yersinebactin (fyuA) and aerobactin (iutA) was 85.6 and 83.0%. Genes encoding capsular antigen synthesis were detected in 61.9% of the strains studied. The kpsMTII and kpsMTIII genes encoding K1, K5, K12 and K3, K10, K54 antigen complexes were detected in 29.4 and 24.2% of the strains. Type-specific gene kpsMT K1, encoding antigen K1, identical in chemical structure and immunochemical characteristics to the K-antigen of Neisseria meningitidis, was detected in 27.3% of strains. The traT gene, a coding factor of bacterial cell resistance to the bactericidal effect of the blood serum, was detected in 59.3% of strains. PAI — UPEC CFT073 was detected in 36.6% of strains.
The genetic determinants of UPEC virulence in the studied strains were present both in combinations and in isolation. None of the tested virulence genes was detected in the genome of 4 (2.1%) strains. In 1 (0.5%; 95% CI 0.1–2.9%) strain 1 gene was detected. The remaining 99.5% of strains were characterized by the presence of gene combinations, of which 1.5% (95% CI 0.5–4.5%) — by the combination of 2 genetic determinants, 10.3% (95% CI 6.8–15.4) — by 3. Strains containing combinations of 4 genes 16.0% (95% CI 11.5–21.8%), 5 genes 23.2% (95% CI 17.8–29.6%), and 6 genes 33.0% (95% CI 26.8–39.9%) were statistically significantly more frequent (p < 0.05). Seven virulence markers were detected in 9.8% (95% CI 6.4–14.8%) of strains, 8 in 3.1% (95% CI 1.4–6.6%), and 9 in 2.1% (95% CI 0.8–5.2%). The genome of 1 strain (0.5%; 95% CI 0.1–2.9%) presented an individual profile and was characterized by a combination of 10 genes.
The occurrence of genes and virulence factors encoded by them in E. coli of different phylogenetic groups are presented in the Table. The fimH gene was present in strains of all phylogenetic groups significantly more frequently (p < 0.05) compared to other genes associated with adhesion. The pap gene was found in E. coli strains of phylogenetic group B2 (36.6%), which was significantly more frequent (p < 0.05) compared to strains of groups A (11.1%), B1 (14.3%) and D (1.7%). The afa gene encoding afimbrial antigen-binding adhesins was more frequently present in strains of phylogenetic group B1 (64.3%). The sfa gene encoding fimbrial adhesin synthesis was detected without significant differences in strains of phylogenetic groups B1 (4.8%) and B2 (11.8%). The focG gene encoding the fimbrial adhesin F1C of uropathogenic E. coli was detected only in strains of phylogenetic group B2 (4.5%).
Occurrence of genes and virulence factors in E. coli strains of various phylogenetic groups
Genes and virulence factors | Phylogenetic groups | |||||||||||
А (n = 9) | В1 (n = 14) | В2 (n = 112) | D (n = 59) | |||||||||
abs. | % | 95% CI | abs. | % | 95% CI | abs. | % | 95% CI | abs. | % | 95% CI | |
Adhesins | 7 | 77,8 | 45,3–93,7 | 14 | 100,0 | 78,5–100 | 110 | 98,2 | 93,7–99,5 | 27 | 45,8 | 33,7–58,7 |
fimH | 5 | 55,6 | 26,7–81,1 | 14 | 100,0 | 78,5–100 | 104 | 92,9 | 86,5–96,3 | 27 | 45,8 | 33,7–58,7 |
pap | 1 | 11,1 | 2,0–43,5 | 2 | 14,3 | 4,0–40,0 | 41 | 36,6 | 28,3–45,8 | 1 | 1,7 | 0,03–9,0 |
afa | 2 | 22,2 | 6,3–54,7 | 9 | 64,3 | 38,8–83,7 | 2 | 1,8 | 0,5–6,3 | 0 | 0 | 0–6,1 |
sfa | 0 | 0,0 | 0–29,9 | 0 | 0,0 | 0–21,5 | 14 | 12,5 | 7,6–19,9 | 1 | 1,7 | 0,03–9,0 |
focG | 0 | 0,0 | 0–29,9 | 0 | 0,0 | 0–21,5 | 1 | 4,5 | 0,2–4,9 | 0 | 0 | 0–6,1 |
Toxins | 3 | 33,3 | 12,1–64,6 | 3 | 21,4 | 7,6–47,6 | 50 | 46,4 | 35,8–53,9 | 6 | 10,2 | 4,8–20,5 |
hlyА | 0 | 0,0 | 0–29,9 | 1 | 7,1 | 1,3–31,5 | 28 | 25,0 | 17,9–33,8 | 3 | 5,1 | 1,7–13,9 |
cnf | 2 | 22,2 | 6,3–54,7 | 0 | 0,0 | 0–21,5 | 22 | 19,6 | 13,3–27,9 | 0 | 0 | 0–6,1 |
cvaС | 1 | 11,1 | 2,0–43,5 | 2 | 14,3 | 4,01–39,95 | 9 | 8,0 | 4,3–14,6 | 4 | 6,8 | 2,7–16,2 |
Invasins | 0 | 0,0 | 0–29,9 | 0 | 0,0 | 0–21,53 | 3 | 2,7 | 0,92–7,58 | 0 | 0 | 0–6,11 |
ibeA | 0 | 0,0 | 0–29,9 | 0 | 0,0 | 0–21,53 | 3 | 2,7 | 0,92–7,58 | 0 | 0 | 0–6,11 |
Siderophore | 3 | 33,3 | 12,1–64,6 | 14 | 100,0 | 78,5–100 | 110 | 98,2 | 93,7–99,5 | 28 | 47,5 | 35,3–59,9 |
fyuA | 3 | 33,3 | 12,1–64,6 | 11 | 78,6 | 52,4–92,4 | 98 | 87,5 | 80,1–92,4 | 23 | 39,0 | 27,6–51,7 |
iutA | 2 | 22,2 | 6,3–54,7 | 11 | 78,6 | 52,4–92,4 | 91 | 81,3 | 73,0–87,4 | 20 | 33,9 | 23,1–46,6 |
Capsules | 1 | 11,1 | 2,0–43,5 | 6 | 42,9 | 21,4–67,4 | 63 | 56,3 | 47,0–65,1 | 9 | 15,3 | 8,2–56,5 |
kpsMTIII | 1 | 11,1 | 2,0–43,5 | 4 | 28,6 | 11,7–54,7 | 26 | 23,2 | 16,4–31,8 | 0 | 0 | 0–6,1 |
kpsMT K1 | 0 | 0,0 | 0–29,9 | 0 | 0,0 | 0–21,5 | 21 | 28,6 | 12,6–27,0 | 3 | 5,1 | 1,7–13,9 |
kpsMTII | 0 | 0,0 | 0–29,9 | 4 | 28,6 | 11,7–54,7 | 31 | 27,7 | 20,2–36,6 | 6 | 10,2 | 4,6–20,5 |
Others | ||||||||||||
traT | 6 | 66,7 | 35,4–87,9 | 10 | 71,4 | 45,4–88,3 | 21 | 18,8 | 12,6–27,0 | 48 | 81,4 | 69,6–89,3 |
PAI | 1 | 11,1 | 2,0–43,5 | 7 | 50,0 | 26,8–73,2 | 37 | 33,0 | 25,0–45,2 | 49 | 83,1 | 71,5–90,5 |
Analysis of the occurrence of genes associated with toxin production showed that genetic markers of toxin formation were statistically significantly more frequent (p < 0.05) in strains of phylogenetic group B2 (46.4%) compared to strains of other phylogenetic groups.
Genetic markers responsible for synthesis of siderophores were present in strains belonging to different phylogenetic groups without significant differences.
Genes encoding the synthesis of K-antigens (kpsMTIII, kps K1, kpsMTII) protecting the bacterial cell from phagocytosis were statistically significantly (p < 0.05) more frequently (56.3%) present in strains of phylogenetic group B2 compared to strains of other phylogenetic groups.
The traT gene and pathogenicity islands (PAI) were found in comparable proportions in strains of all phylogenetic groups.
Discussion
The main difficulty in interpreting the results of the culture method in the detection of extraintestinal pathogenic E. coli (ExPEC) strains is presented by isolates from urine specimens, due to the lack of clearly defined criteria for assessing the etiological significance of a particular isolate, as well as due to possible contamination of the sample. True ExPEC pathogens, including UPEC, include strains containing 2 or more major virulence genes (pap, sfa, afa, kpsMTII, iutA). Other so-called additional genes (fimH, hlyA, cvaC, cnf, cdtB, kpsMTIII, ibeA, traT and PAI) may be potentially associated with ExPEC as they promote adaptive and competitive colonization [17, 18]. E. coli strains that, in addition to the main ExPEC virulence genes, have potential ones, are characterized by an increased ability to adapt to new niches, which allows them to cause a wide range of diseases.
The E. coli strains isolated from urine belonged to different phylogenetic groups, with the majority (88.1%) belonging to groups B2 (57.7%) and D (30.4%), which are usually associated with pathogens of extraintestinal localization [19]. Groups A and B1, with which commensal E. coli are associated, were assigned to 4.6 and 7.2% of strains [16]. Pathogenetically significant genetic determinants of virulence were identified in 97.9% of strains. According to the combination of 17 genes associated with adhesion (fimH, pap, afa, sfa and focG), synthesis of siderophores (fyuA and iutA), capsules (kpsMTII, kpsMTIII and kpsMT K1), toxins (hlyA, cnf, cdt and cvaC), invasins (ibeA), providing serum resistance (traT), presence of pathogenicity islands (PAI), 134 individual virulence genotypes were identified. The vast majority (99.5%) of strains contained between 2 and 10 virulence genes. The results obtained about the pronounced genetic heterogeneity of UPEC strains are consistent with the data of Russian researchers [20, 21]. The study showed that 95.9% (186/194) of strains isolated from urine met the UPEC criteria — they had 3–10 genes, belonged to phylogroups B2 and D. Eight (4.1%) strains belonged to phylogroups A and B1, had no major or additional virulence genes and were considered as contaminants.
Genetic predisposition to UTI recurrence was detected in 93.3% of strains due to the presence of fimH adhesins encoded by the fimH gene, which promote UPEC persistence [5–7]. In addition to fimbrial adhesins, afa-encoded adhesins of the Afa/Dr family, which promote adhesion with urothelial cells, are widely distributed in UPEC. In this study, strains synthesizing afibrial adhesins, which, according to other researchers, have a high potential to cause pyelonephritis and recurrent cystitis, were identified in 13 patients examined in this study [22].
Meningitis-associated E. coli (NMEC) is a common cause of neonatal bacterial meningitis with a mortality rate of up to 40% and severe neurologic sequelae [23, 24]. Most cases of neonatal infection occur when NMEC are present in the urogenital tract of parturients, carried in the intestine, or as a complication of neonatal sepsis [25]. Escherichia coli sepsis is often considered as a secondary infection. One in three cases is urosepsis, which can occur in any urologic disease (urodynamic disorders in UTIs, purulent forms of pyelonephritis, urinary retention, acute prostatitis) [26]. The risk of sepsis development is increased in the infectious process caused by E. coli strains that contain P-pili and S-fimbriae, producing hemolysin, cytonecrotic factor and synthesizing capsular antigen K2 [8, 14]. Markers of unfavorable prognosis of UTI course were found in 12% of strains, 10.7% of which had genetic determinants associated with the development of sepsis (urosepsis), 1.3% with meningitis.
The presence of rather contradictory results in the application of the traditional bacteriological method of laboratory research makes us critically reevaluate its diagnostic significance in favor of the undoubtedly promising molecular genetic method. The generally accepted practice of unconditionally recognizing the etiological significance of a specific isolate that is quantitatively predominant in a sample during culture looks at least questionable [27]. Nevertheless, it is impossible to abandon the generally accepted method of culturing microorganisms due to the fact that it allows obtaining information about clinically significant strains, their number and associations, assessing sensitivity to antibiotics, bacteriophages and disinfectants, as well as being an integral step in DNA isolation for subsequent molecular studies, including whole genome sequencing.
Conclusion
The study showed that detection of the gene complex in E. coli strains isolated from urine not only confirms the etiologic significance of the isolate, but also makes it possible to assess the pathogenic potential for the development of chronic forms and acute life-threatening complications. The revealed heterogeneity of the UPEC population indicates the necessity to optimize the algorithm and develop standards for laboratory diagnosis and prevention of complications of diseases caused by uropathogenic E. coli. The results of the integrative approach in the laboratory diagnosis of UTI using all adequate methods (traditional, modern, innovative), centered on the principles of evidence-based medicine, in other words, reliability of scientific evidence of their effectiveness, should be taken into account in the practice of specialists of different profiles.
About the authors
Mariia A. Makarova
Saint-Peterburg Pasteur Institute; North-Western State Medical University named after I.I. Mechnikov
Author for correspondence.
Email: makmaria@mail.ru
ORCID iD: 0000-0003-3600-2377
D. Sci. (Med.), senior researcher, Head, Laboratory of enteric infection, Saint-Peterburg Pasteur Institute, St. Petersburg, Russia; assistant professor, Department of medical microbiology, North-Western State Medical University named after I.I. Mechnikov
Russian Federation, St. Petersburg; St. PetersburgZoya N. Matveeva
Saint-Peterburg Pasteur Institute
Email: makmaria@mail.ru
ORCID iD: 0000-0002-7173-2255
Cand. Sci. (Med.), senior researcher, Laboratory of enteric infection, Saint-Peterburg Pasteur Institute
Russian Federation, St. PetersburgLidiya A. Kaftyreva
Saint-Peterburg Pasteur Institute; North-Western State Medical University named after I.I. Mechnikov
Email: makmaria@mail.ru
ORCID iD: 0000-0003-0989-1404
D. Sci. (Med.), senior researcher, Head, Microbiology department, Saint-Peterburg Pasteur Institute; assistant professor, Department of medical microbiology, North-Western State Medical University named after I.I. Mechnikov
Russian Federation, St. Petersburg; St. PetersburgReferences
- Бухарин О.В., Гриценко В.А., Вялкова А.А. Факторы уропатогенности бактерий: роль в патогенезе и значение в диагностике пиелонефрита. Нефрология и диализ. 2001;3(4): 469–75. Bukharin O.V., Gritsenko V.A., Vyalkova A.A. Factors of uropathogenicity of bacteria: role in pathogenesis and significance in the diagnosis of pyelonephritis. Nephrology and Dialysis. 2001;3(4):469–75. EDN: https://elibrary.ru/wjcffv
- Ali I., Rafaque Z., Ahmed I., et al. Phylogeny, sequence-typing and virulence profile of uropathogenic Escherichia coli (UPEC) strains from Pakistan. BMC Infect. Dis. 2019;19(1):620. DOI: https://doi.org/10.1186/s12879-019-4258-y
- European Association of Urology. Guidelines on urological infection; 2018. Available at: https://uroweb.org/wp-content/uploads/EAU-Guidelines-on-Urological-Infections-2018-large-text.pdf
- Кузнецова М.В., Гизатуллина Ю.С. Генетические профили адгезии и адгезивная вариабельность уропатогенных штаммов Escherichia coli. Инфекция и иммунитет. 2021;11(3):481–90. Kuznetsova M.V., Gizatullina J.S. Genetic adhesion profiles and adhesive variability of uropathogenic Escherichia coli strains. Russian Journal of Infection and Immunity. 2021;11(3):481–90. DOI: https://doi.org/10.15789/2220-7619-GAP-1413 EDN: https://elibrary.ru/edkmlc
- Hancock S.J., Lo A.W., Ve T., et al. Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli. PLoS Pathog. 2022;18(6):e1010582. DOI: https://doi.org/10.1371/journal.ppat.1010582
- Etefia E.U., Ben S.A. Virulence markers, phylogenetic evolution, and molecular techniques of uropathogenic Escherichia coli. J. Nat. Sci. Med. 2020;3(1):13–22. DOI: https://doi.org/10.4103/JNSM.JNSM_31_19
- Захарова И.Н., Османов И.М., Мачнева Е.Б. и др. От бактериурии до микробиома мочевых путей: эволюция взглядов ученых и клиницистов. Медицинский совет. 2018;(17):168–77. Zakharova I.N., Osmanov I.M., Machneva E.В., et al. From bacteriuria to the urinary tract microbiome: the evolution of the views of researchers and clinicians. Medical Council. 2018;(17):168–77. DOI: https://doi.org/10.21518/2079-701X-2018-17-168-176 EDN: https://elibrary.ru/ylhwzv
- Sarowska J., Futoma-Koloch B., Jama-Kmiecik A., et al. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 2019;11:10. DOI: https://doi.org/10.1186/s13099-019-0290-0
- Meza-Segura M., Zaidi M.B., Maldonado-Puga S., et al. Cytolethal distending toxin-producing Escherichia coli strains causing severe diarrhoea in young Mexican children. JMM Case Rep. 2017;4(2):e005079. DOI: https://doi.org/10.1099/jmmcr.0.005079
- Starčič Erjavec M., Žgur-Bertok D. Virulence potential for extraintestinal infections among commensal Escherichia coli isolated from healthy humans – the Trojan horse within our gut. FEMS Microbiol. Lett. 2015;362(5):fnu061. DOI: https://doi.org/10.1093/femsle/fnu061
- Banerjee R., Weisenhorn E., Schwartz K.J., et al. Tailoring a global iron regulon to a uropathogen. mBio. 2020;11(2):e00351-20. DOI: https://doi.org/10.1128/mbio.00351-20
- Камалов А.А., Ходырева Л.А., Дударева А.А., Низов А.Н. Факторы риска развития инфекционно-воспалительного процесса нижних мочевых путей. Вестник дерматологии и венерологии. 2015;91(2):63–7. Kamalov A.A., Khodyreva L.A., Dudareva A.A., Nizov A.N. Risk factors causing the development of infection and inflammation of the lower urinary tract. Bulletin of Dermatology and Venereology. 2015;91(2):63–7. DOI: https://doi.org/10.25208/0042-4609-2015-91-2-63-67 EDN: https://elibrary.ru/tyjeip
- Abe C.M., Salvador F.A., Falsetti I.N., et al. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol. Med. Microbiol. 2008;52(3):397–406. DOI: https://doi.org/10.1111/j.1574-695x.2008.00388.x
- Daga A.P., Koga V.L., Soncini J.G.M., et al. Escherichia coli bloodstream infections in patients at a University Hospital: virulence factors and clinical characteristics. Front. Cell. Infect. Microbiol. 2019;9:191. DOI: https://doi.org/10.3389/fcimb.2019.00191.
- Nojoomi F., Ghasemian A. The relation of phylogroups, serogroups, virulence factors and resistance pattern of Escherichia coli isolated from children with septicemia. New Microbes New Infect. 2019;29:100517. DOI: https://doi.org/10.1016/j.nmni.2019.100517
- Clermont O., Bonacorsi S., Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000;66(10):4555–8. DOI: https://doi.org/10.1128/aem.66.10.4555-4558.2000
- Johnson J.R., Russo T.A. Molecular epidemiology of extraintestinal pathogenic Escherichia coli. EcoSal Plus. 2018;8(1). DOI: https://doi.org/10.1128/ecosalplus.esp-0004-2017
- Russo T.A., Johnson J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 2003;5(5): 449–56. DOI: https://doi.org/10.1016/s1286-4579(03)00049-2
- Hernández-Chiñas U., Ahumada-Cota R.E., Navarro-Ocaña A., et al. Phenotypic and genotypic characteristics of Escherichia coli strains isolated during a longitudinal follow-up study of chronic urinary tract infections. Front. Public Health. 2023;11:1240392. DOI: https://doi.org/10.3389/fpubh.2023.1240392
- Казанцев А.В., Осина Н.А., Глинская Т.О. и др. Факторы вирулентности и филогенетическая характеристика уропатогенных штаммов Eschericihia coli, выделенных на территории г. Саратова. Проблемы особо опасных инфекций. 2019;(4):56–60. Kazantsev A.V., Osina N.A., Glinskaya T.O., et al. Virulence factors and phylogenetic characteristics of uropathogenic Eschericihia coli strains isolated in Saratov. Problems of Particularly Dangerous Infections. 2019;(4):56–60. DOI: https://doi.org/10.21055/0370-1069-2019-4-56-60 EDN: https://elibrary.ru/gplihe
- Слукин П.В., Асташкин Е.И., Асланян Е.М. и др. Характеристика вирулентных штаммов Escherichia coli, выделенных от пациентов с урологической инфекцией. Журнал микробиологии, эпидемиологии и иммунобиологии. 2021;98(6):671–84. Slukin P.V., Astashkin E.I., Aslanyan E.M., et al. Characterization of virulent Escherichia coli strains isolated from patients with urological infection. Journal of Microbiology, Epidemiology and Immunobiology. 2021;98(6):671–84. DOI: https://doi.org/10.36233/0372-9311-134 EDN: https://elibrary.ru/rquaau
- Whelan S., Lucey B., Finn K. Uropathogenic Escherichia coli (UPEC)-associated urinary tract infections: the molecular basis for challenges to effective treatment. Microorganisms. 2023;11(9):2169. DOI: https://doi.org/10.3390/microorganisms11092169
- Zhu N., Liu W., Prakash A., et al. Targeting E. coli invasion of the blood-brain barrier for investigating the pathogenesis and therapeutic development of E. coli meningitis. Cell Microbiol. 2020;22(10):e13231. DOI: https://doi.org/10.1111/cmi.13231
- Rudd K.E., Johnson S.C., Agesa K.M., et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395(10219):200–11. DOI: https://doi.org/10.1016/s0140-6736(19)32989-7
- Багирова Н.С. Бактериемия истинная или ложная: значение критериев оценки клинической значимости положительной гемокультуры. Клиническая лабораторная диагностика. 2015;60(8):55–61. Bagirova N.S. The true or false bacteriemia: the significance of evaluation criteria of clinical significance of positive hemoculture. Clinical Laboratory Diagnostics. 2015;60(8):55–61. EDN: https://elibrary.ru/uiqjnx
- Мельников В.Л., Митрофанова Н.Н., Суменкова А.О., Терина Н.А. Гнойно-септические осложнения в урологическом отделении стационара (обзор литературы). Известия высших учебных заведений. Поволжский регион. Медицинские науки. 2019;(3):39–53. Mel'nikov V.L., Mitrofanova N.N., Sumenkova A.O., Terina N.A. Purulent-septic complications in the urology unit (literature review). University Proceedings. Volga Region. Medical Sciences. 2019;(3):39–53. DOI: https://doi.org/10.21685/2072-3032-2019-3-4 EDN: https://elibrary.ru/wkgtvt
- Годовалов А.П., Николаева Н.В., Карпунина Т.И., Оборин Д.А. К оценке этиологической значимости бактерий, детектированных в генитальном тракте мужчин. Журнал микробиологии, эпидемиологии и иммунобиологии. 2022;99(4): 428–35. Godovalov A.P., Nikolaeva N.V., Karpunina T.I., Oborin D.A. On the assessment of the etiological significance of bacteria detected in the male genital tract. Journal of Microbiology, Epidemiology and Immunobiology. 2022;99(4):428–35. DOI: https://doi.org/10.36233/0372-9311-257 EDN: https://elibrary.ru/npmvrq
Supplementary files