Single intranasal immunization with attenuated Wuhan-like SARS-CoV-2 provides highly effective cross-protection against Delta and Omicron variants of concern
- Authors: Faizuloev E.B.1,2, Gracheva A.V.1, Korchevaya E.R.1, Ammour Y.I.1, Smirnova D.I.1, Khokhlova D.M.1, Drokov A.O.3, Pankratov A.A.4, Trunova G.V.4, Khokhlova V.A.4, Vorontsova M.S.4, Leneva I.A.1, Svitich O.A.1,3, Zverev V.V.1,3
-
Affiliations:
- I.I. Mechnikov Research Institute of Vaccines and Sera
- Russian Medical Academy of Continuous Professional Education
- I.M. Sechenov First Moscow State Medical University (Sechenov University)
- P.A. Hertsen Moscow Oncology Research Institute
- Issue: Vol 101, No 1 (2024)
- Pages: 36-51
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18552
- DOI: https://doi.org/10.36233/0372-9311-496
- EDN: https://elibrary.ru/zqbojh
- ID: 18552
Cite item
Abstract
Introduction. Despite the end of the COVID-19 pandemic, the problem of vaccine prevention of this disease appears highly relevant. The emergence and widespread distribution of the Omicron SARS-CoV-2 variant of concern (VOC) and its sublineages has dramatically reduced the efficacy of vaccination. The possible approach to solving this problem is to develop a nasal live attenuated vaccine capable of activating humoral, mucosal, and cell-mediated immunity, providing a prolonged immune response and cross-protection against different VOCs.
The aim of the study was to determine the immunization efficacy with attenuated cold-adapted Wuhan-like SARS-CoV-2 D-D2 strain against homologous and heterologous challenges.
Materials and methods. The study was conducted on an animal model of coronavirus pneumonia in golden Syrian hamsters. The efficacy of immunization was assessed by comparing the dynamics of weight, viral load in organs and histopathological changes in the lungs in immunized and unimmunized animals.
Results. Single intranasal immunization of golden Syrian hamsters with D-D2 strain showed its high immunogenicity: seroconversion was evident in all immunized animals. Wuhan-like D-D2 strain provides highly effective protection of hamsters against the development of productive infection and pneumonia when challenged both with ancestral virus and heterologous strains related to Delta (AY.122) and Omicron (sublineages BA.1.1 and BA.5.2) variants.
Conclusion. SARS-CoV-2 attenuation is a promising strategy for the development of a highly effective nasal live COVID-19 vaccine.
Full Text
Introduction
Despite the end of the COVID-19 pandemic declared by WHO, the problem of vaccine prevention of this disease continues to be relevant. Inactivated, vector, subunit, and mRNA vaccines are used for COVID-19 prevention and have been shown to be highly effective against homologous infection (i.e., infection with the same virus variant as the strain used for vaccine) [1]. The emergence and widespread distribution of the SARS-CoV-2 Omicron variant of concern (VOC) and its sublineages have dramatically reduced the efficacy of vaccination [1–3]. Most licensed COVID-19 vaccines are directed toward the formation of humoral immunity based on the induction of neutralizing antibodies to the SARS-CoV-2 S-protein. However, the evolution of the virus under the mass immunization pressure makes this target highly variable, resulting in the virus escaping immunological surveillance and a rapid decline in vaccine efficacy against newly emerging SARS-CoV-2 variants [2–4].
Emergent optimization of the composition of existing vaccines according to the current set of circulating SARS-CoV-2 variants is necessary to maintain vaccination efficacy at a high level [5]. Development of a "universal" vaccine with cross-protective activity against different antigenic variants of the virus could represent an alternative approach. Such approach includes development of a live attenuated vaccine (LAV) capable of activating not only the humoral but also the cellular immune system, providing a prolonged immune response and cross-protection against different virus variants [6–8]. At present, the potential of LAV in COVID-19 prevention remains unrealized. At the same time, some experience has already been gained in obtaining attenuated SARS-CoV-2 strains that have shown high immunogenicity and immunization efficacy in animal models of infection. Some authors use genetic engineering and reverse genetics techniques such as site-directed mutagenesis and codon deoptimization to attenuate the virus [9–13]. Others use the traditional approach to obtain virus mutants by prolonged passaging in cell culture under selective conditions [14–17]. Regardless of the methodology used to attenuate the virus, an important question that determines the practical relevance of LAV at the current stage is whether it can provide effective protection against the "parent" SARS-CoV-2 strain, but also against new, phylogenetically distant virus variants.
Earlier, by long-term passaging of Wuhan-like SARS-CoV-2 Dubrovka strain in Vero cells at lower temperature (up to 23ºC), we obtained its cold-adapted (ca) variant called D-D2 strain [18]. D-D2 strain exhibited a temperature-sensitive (ts) phenotype (it did not replicate at 39ºC), which determined its reduced replication in the lungs of golden Syrian hamsters and, thus, an attenuated (att) phenotype. During intranasal immunization of hamsters, D-D2 strain exhibited high immunogenicity and protected against infection with homologous parental Dubrovka strain and the development of pneumonia [15]. The aim of the present study was to determine the immunization efficacy of attenuated Wuhan-like SARS-CoV-2 strain against heterologous challenge with strains belonging to the Delta variant and two sublineages of Omicron variant, BA.1.1 and BA.5.2. For immunization, ca/ts/att D-D2 strain was used as a model vaccine strain.
Materials and methods
Viruses. To model coronavirus pneumonia on Syrian golden hamsters (hereinafter referred to as hamsters) we used laboratory strains of SARS-CoV-2 (family: Coronaviridae, genus: Betacoronavirus, subgenus: Sarbecovirus, species: Severe acute respiratory syndrome–related coronavirus): Dubrovka (Wuhan-like), Podolsk (Delta); Otradnoe (Omicron BA.1.1), and FEB2 (Omicron BA.5.2) isolated and genetically characterized in different periods of pandemic in our laboratory (Table 1). A cold-adapted D-D2 strain with ts and att phenotype, which we obtained earlier by adapting the Dubrovka strain for growth in Vero CCL-81 cell culture at lower temperature (23ºC), was used to immunize animals [18].
Table 1. SARS-CoV-2 strains used in the study
Strain | Collection date | GenBank ID | Variant | Pangolin lineage | Passage level | Cultivation temperature, ºC |
Dubrovka | June 2020 | MW514307.1 | Wuhan-like (wild-type) | B.1.1.317 | 17 | 37 |
D-D2 | June 2020 | ON040961.1 | Wuhan-like (cold-adapted) | B.1.1.317 | 47 | 23 |
Podolsk | August 2021 | ON032860.1 | Delta | AY.122 | 16 | 37 |
Otradnoe | January 2022 | ON032857.1 | Omicron | BA.1.1 | 8 | 37 |
FEB2 | October 2022 | OP920753.1 | Omicron | BA.5.2 | 4 | 37 |
Virus cultivation in cell culture
SARS-CoV-2 laboratory strains were cultured on African green monkey kidney epithelial Vero CCL-81 cells (ATCC), as described earlier [19]. Monolayer of Vero cells was infected with the SARS-CoV-2 at a low multiplicity of infection (MOI ≤ 0,001) and incubated at 37ºC (Dubrovka, Podolsk, Otradnoe and FEB2 strains) or 23ºC (D-D2 strain) for 3-8 days (depending on virus strain) in an atmosphere of 5% CO2. Virus-containing culture medium was clarified by centrifugation and stored at –80ºC before use.
Animals
Four-week-old 40–50 g female Syrian golden hamsters (Mesocricetus auratus) were obtained from the Nursery for laboratory animals of the Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry (Russia). Hamsters were arbitrarily assigned to study groups. Animals were kept as described earlier [20]. All studies with animals were approved by the Mechnikov Research Institute of Vaccines and Sera Institutional Animal Care and Use Committee (protocol No. 2, May 24, 2021) and carried out in accordance with the National Research Council's Guide for the Care and Use of Laboratory Animals (2011).
Virus titration
The SARS-CoV-2 virus titers were determined by the cytopathic effect endpoint method in Vero cells as described earlier [19]. The virus titer was calculated by the Reed–Muench method using an online-calculator1 and expressed as log10 TCID50/ml.
Quantification of SARS-CoV-2 RNA
Quantification of SARS-CoV-2 RNA was performed by real-time reverse transcription polymerase chain reaction (RT-PCR) method [20]. Viral RNA was isolated from samples using “MagnoPrime UNI” reagent kit (“NextBio”), recommended by the manufacturer for isolation viral RNA from a wide range of biological and clinical samples. To detect viral RNA we used primers and the probe designed for the SARS-CoV-2 nucleocapsid N gene: CoVN-F GCGTTCTTCGGAATGTCG, COVN-R TTGGATCTTTGTCATCCAATTTG, COVN-P FAM-AACGTGGTTGACCTACACAGGT-BHQ1 [21]. A 2.5x Taq-polymerase reaction mixture reagent kit and MMLV reverse transcriptase (Syntol) were used to perform one-step reverse transcription real-time PCR reaction. 50 µL reaction mixture contained 5 units of Taq DNA polymerase, 30 units of MMLV reverse transcriptase, 10 pmol of each primer and 5 pmol of probe. Thermal cycling process included 45ºC — 10 min (1 cycle); 95ºC — 5 min (1 cycle); 95ºC — 5 sec, 55ºC — 45 sec (45 cycles). Synthetic oligonucleotide corresponding to the fragment of SARS-CoV-2 genome containing the primers and probe binding sites were used to construct a calibration curve: COVN-PC CAGCGTTCTTCGGAATGTCGCGCATTGGCATGGAAGTCACACCTTCGGGAACGTGGTTGACCTACACAGGTGCCATCAAATTGGATGACAAAGATCCAAA. The analytical sensitivity of the real-time PCR established by analyzing 10-fold dilutions of the COVN-PC oligonucleotide (SARS-CoV-2 cDNA standard) was 5 × 102 copies per ml. Adjusted for losses at the stages of RNA isolation and reverse transcription, the sensitivity of SARS-CoV-2 RNA detection was assumed to be 103 copies per ml. The typical standard curve of dependence of Ct values on the concentration of viral RNA was described by the equation:
Y = 48.816 × 3.5348X,
where Y is Ct value; X is concentration of viral RNA (Log10 copies/mL).
The results of RT-PCR were expressed as copies of viral RNA per mL of tissue homogenate.
Evaluation of the immunization efficacy
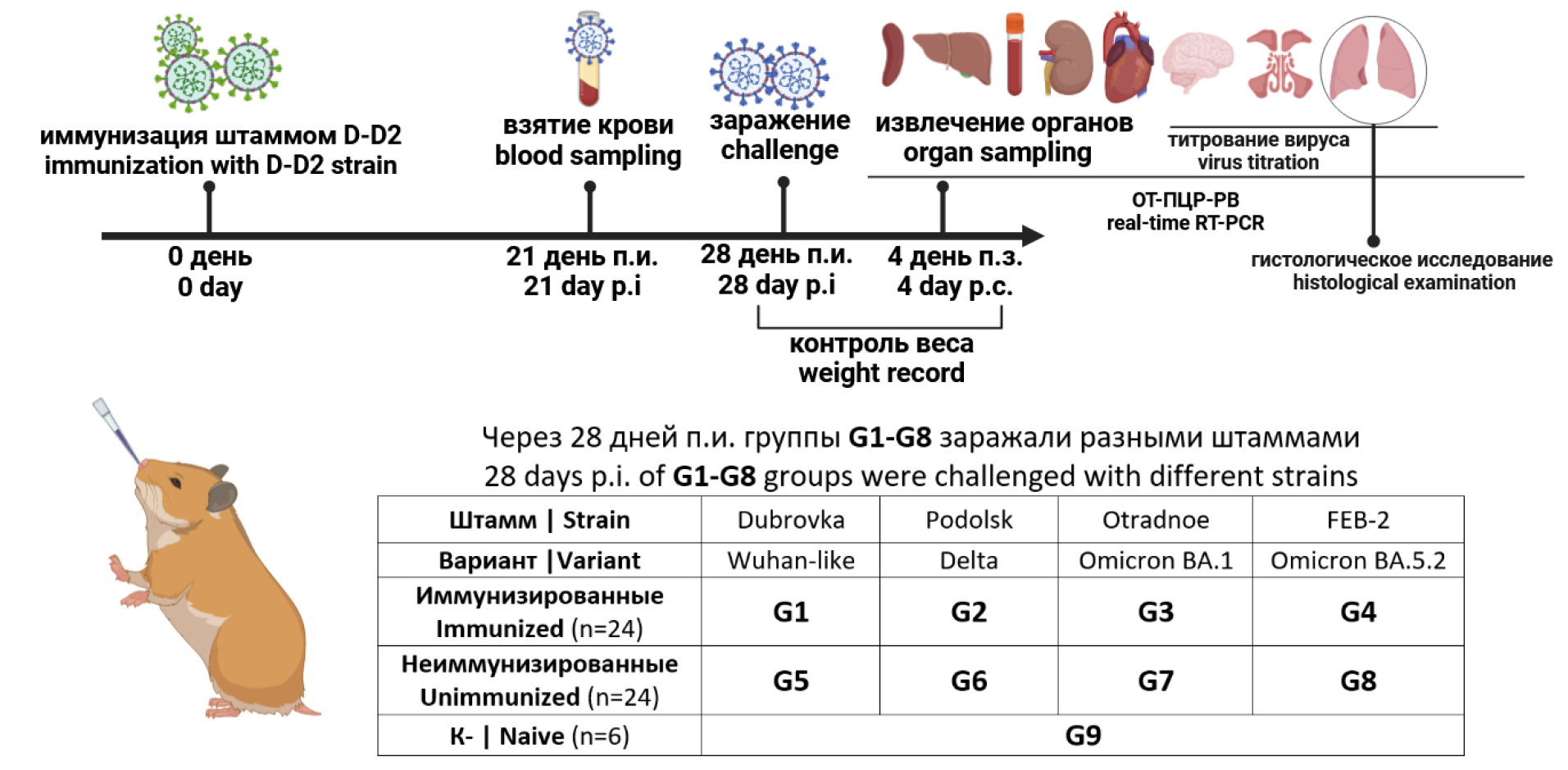
Immunization efficacy was assessed according to the scheme (Fig. 1). Syrian hamsters were divided into nine groups (G1–G9) of six animals each. Each animal of groups G1–G4 received intranasally 4.0 log10 TCID50 in 100 μL of the D-D2 strain. Intranasal administration of the virus was carried out under diethyl ether anesthesia. Animals of groups G5–G9 were not immunized and received PBS intranasally. 21 days post-immunization (p.i.), blood was taken from the animals to assess the total and neutralizing antibodies to SARS-CoV-2. To evaluate the immunization efficacy 28 days p.i., each animal of groups 1-8 was challenged by 4.0 log10 TCID50 in 100 μL of the virulent SARS-CoV-2 strain intranasally. Groups G1 and G5 received ancestral Dubrovka strain, groups G2 and G6 — Podolsk strain (Delta), groups G3 and G7 — Otradnoe strain (Omicron BA.1.1), groups G4 and G8 — FEB2 strain (Omicron BA.5.2). The negative control group G9 received an equivalent amount of PBS. Hamsters were observed daily and weight control was performed daily from days 0 to 4. Four days after the challenge, animals were humanely euthanized under chloroform anesthesia. Left lungs were collected for histological examination. Lung, brain, and other organ tissues were collected, homogenized in DMEM medium with gentamicin (40 µg/mL) using a “Tissue Lyser LT homogenizer” (“Qiagen”), and centrifuged at 10,000 rpm for 5 min at 4ºC. Supernatants were collected and stored at –80ºC for subsequent titration and determination of viral RNA. The immunization efficacy was evaluated by comparing the weight dynamics, viral load in the organs, and histopathological changes in lungs in immunized and unimmunized animals.
Fig. 1. The design of the experiment for evaluating the immunization efficacy.
Lung histological examination
Lung histological examination was performed as described earlier [15]. Formalin-fixed paraffin-embedded left hamster lungs stained by hematoxylin and eosin were used for histological study. Histological sections of the lungs scored blindly for lung damage, using a cumulative severity score of 0 to 3 each for an extended range of parameters as recently suggested [22], i.e. % of lung area affected, distribution of lesions, necrosis of bronchial ciliated epithelial cells, cellular debris in bronchi, diffuse alveolar damage, necrosis of alveolar cells, cellular debris in alveoli, alveolar hemorrhage, alveolar edema, perivascular/interstitial edema, vasculitis, necrosis and desquamation of vascular endothelial cells, bronchitis, bronchointerstitial pneumonia, interstitial pneumonia, intraalveolar neutrophils and macrophages, hyperplasia of bronchial ciliated epithelial cells, hyperplasia of type II alveolar cells. The maximum possible score was 60.
Detection of antibodies to SARS-CoV-2
Detection of antibodies to SARS-CoV-2 in hamster’s sera was performed by the ELISA method as described earlier [15]. For ELISA, 96-well plates were coated with a UV-inactivated SARS-CoV-2 virions of Dubrovka strain, prepared as described by [20].
Viral neutralization test
Measurement of SARS-CoV-2 neutralizing antibodies based on cytopathic effect in Vero cells was performed according to the protocol described earlier [19]. Dubrovka, Podolsk, Otradnoe, and FEB2 SARS-CoV-2 strains adapted to Vero cells were used to determine the neutralizing activity against different antigenic variants. The neutralizing titer was considered the reciprocal value of the last dilution, in which no signs of cytopathic effect were detected in two or more wells.
Statistical analysis
Statistical processing was performed using GraphPad Prism v. 5.03 software. The data were presented as the mean ± standard deviation (SD) and mean ± standard error (SEM) on the plots. The differences were compared using paired sample t-test. Differences were considered to be significant at p < 0.05.
Work safety requirements
All work with the SARS-CoV-2 virus was carried out under conditions of a Biosafety Level-3 laboratory.
Results
Evaluation of the immunogenicity and efficacy of attenuated D-D2 strain (a cold-adapted mutant of Dubrovka strain) was performed in an experiment on golden Syrian hamsters immunization according to the scheme (Fig. 1). Hamsters of groups G1–G4 (n = 24) were intranasally immunized with the D-D2 strain at the dose of 4.0 log10 TCID50 per animal.
Immunogenicity evaluation
After 21 days p.i., IgG antibodies to SARS-CoV-2 proteins in ELISA were detected in sera of all immunized animals in titers ranging from 12,800 to 204,800 (mean 54,933 ± 44,257) (Fig. 2, a). The virus-neutralizing activity of hamsters’ sera after immunization with the D-D2 strain was maximal for homologous Dubrovka strain 793 ± 524 (Fig. 2, b). For heterologous strains, the neutralizing activity of sera was lower for the Podolsk strain (Delta) by four times for Otradnoe and FEB2 strains (Omicron) by 46 times. In 56% of sera samples, the neutralizing activity against Otradnoe and FEB2 strains was not detected. In the sera of non-immunized animals (groups G5–G9; n = 30) no antibodies to the virus were detected either by ELISA or in the neutralization test.
Fig. 2. Immunogenicity of the D-D2 strain in hamsters following single intranasal administration. Hamsters of G1–G4 groups were immunized with the D-D2 strain. a — titer of IgG antibody to SARS-CoV-2 proteins by ELISA; b — titers of neutralizing antibodies against different SARS-CoV-2 strains in the sera of all animals from groups G1–G4 (n = 24) — Dubrovka (Wuhan), Podolsk (Delta), Otradnoe (BA.1.1), and FEB2 (BA.5.2). The limit of detection was 20. ***p < 0.001.
Evaluation of the immunization efficacy
Twenty-eight days after immunization, the animals were challenged with different SARS-CoV-2 strains according to the scheme (Fig. 1). Challenge of unimmunized animals (groups G5-G8) with different strains revealed their different virulence for Syrian hamsters. The Dubrovka strain proved to be the most virulent and resulted in weight loss of 14.6% (p = 0.0002) on average at day 4 p.c. (Fig. 3), the most severe pneumonia (Fig. 4, 5), infection not only respiratory tract but also brain (Fig. 6, 7), that was accompanied by decreased appetite, lethargy, and sleepiness. When unimmunized animals were infected with Podolsk, Otradnoe, and FEB2 strains, weight loss was not significant (p > 0.05), average by 2.15%, 1.2%, and 2.4% compared to group G9 (naive), while lung histopathology and the viral load in the brain were lower than infected with the Dubrovka strain (Fig. 4–7).
Fig. 3. Weight change of immunized and unimmunized hamsters at days 1–4 post-challenge.
Fig. 4. Morphological changes of hamster lungs on day 4 post-challenge with different SARS-CoV-2 strains. Hematoxylin and eosin staining. The size of the scale bar is expressed in microns.
Fig. 5. Histopathology score for hamster lungs on day 4 post-challenge with different SARS-CoV-2 strains. *р < 0.05; **р < 0.01; ***р < 0.001.
Fig. 6. Virus titer in lungs, nasal passages, and brain of immunized and unimmunized hamsters. The limit of detection was 2.0 log10 TCID50/mL.
Fig. 7. Concentration of viral RNA in organs of immunized and unimmunized hamsters on 4 days post-challenge. a — lungs, nasal passages, blood, and brain; b — liver, heart, kidneys and spleen. The limit of detection was 3.0 log10 RNA copies/mL. *p < 0.05; **p < 0.01; ***p < 0.001.
All immunized hamsters showed no delay in weight gain and no changes in behavior compared to unimmunized animals (Fig. 3). Among the immunized animals (groups G1–G4), the strongest protection against weight loss was observed in group G1 infected with the Dubrovka strain, reaching 12.7% (p < 0.0001) at day 4 p.c. When infected with the strains Podolsk (group G2) and FEB2 (group G4) the difference in the weight of the immunized and unimmunized animals was non-significant (p > 0.05), reaching 2.61% and 2.48% at day 4 p.c., respectively. When infected with the strain Otradnoe (group G3) the difference in the weight of the immunized and unimmunized animals was significant (p = 0.02), reaching 2.45% at days 3 and 4 p.c.
Morphological changes in the lungs of challenged unimmunized hamsters (groups G5–G9) corresponded to interstitial pneumonia, but the severity and prevalence of the lesions depended on the strain (Fig. 4). On the fourth day post-challenge (p.c.) with Wuhan-like strain Dubrovka (group G5), histological preparations of animal lungs showed pronounced alterative and inflammatory changes, which morphologically corresponded to bronchointerstitial pneumonia. Areas of pneumonia occupied from 50% to 90% of the histological section of the organ indicating the subtotal degree of spread of the inflammatory process in hamster lungs. In the lungs of hamsters of group G6 challenged with the Podolsk (Delta) strain, foci of bronchointerstitial pneumonia occupied from 15 to 50% of the organ slice area. The lung morphology of hamsters in groups 7 and 8, challenged with the Otradnoe (Omicron BA 1.15) and FEB (Omicron BA 5.2) strains, showed significant differences, despite the close phylogenetic relationship between these strains. After the challenge with the Otradnoe strain, the lungs preparations revealed foci of bronchointerstitial pneumonia, which occupied not more than 5–7% of the organ slice area. Contrary, when infected with the FEB2 strain, foci of pneumonia occupied from 40 to 60%, and the morphological pattern of inflammatory changes was similar to that in hamsters challenged with the Dubrovka strain (Fig. 4).
Histological examination of the lungs of immunized hamsters (groups G1–G4) 4 days p.c. with different strains did not reveal pathological changes, while inflammatory changes were mild (Fig. 4). No pathological changes were detected in hamster lungs immunized and challenged with Dubrovka (group G1) and Podolsk (group G2) strains. The histological structure of the airways and respiratory zone of the lungs appeared normal. Lungs from immunized hamsters challenged with Otradnoe (group G3) and FEB2 (group G4) strains showed focal slightly expressed inflammatory changes in the bronchi, while the morphology of respiratory zone did not differ from those in negative control animals (group G9).
Histological examination of lung preparations of unimmunized uninfected hamsters (group G9, naive) revealed no pathological changes (Fig. 4).
Thus, histological examination of the lungs showed that immunization with the D-D2 strain protects animals from the development of viral pneumonia regardless of the SARS-CoV-2 strain used for infection.
Based on histological examination, pathological changes in the lungs of animals of groups G1–G9 were given a score according to A.D. Gruber et al. [22] (Fig. 5). In immunized animals (groups G1–G4), cumulative severity score varied on average from 3.2 to 6.9, while in unimmunized animals (groups G5–G9) — from 20.8 to 49.8. Immunization of hamsters with the D-D2 strain reduces the lungs histopathology score when challenged with the Dubrovka strain by 15.7 times (p < 0.01), Podolsk — 8.4 times (p < 0.01), Otradnoe — 4.0 times (p < 0.01), FEB2 — 5.7 times (p < 0.01). At the same time, immunized animals challenged with Omicron-like strains Otradnoe and FEB2 had significantly higher lungs histopathology scores (5.2 ± 1.5 and 6.9 ± 0.9, respectively), compared to hamsters challenged with ancestral Dubrovka strain (3.2 ± 0.1).
Single intranasal immunization with the D-D2 strain protected hamsters from developing a productive infection when infected with all SARS-CoV-2 strains as evidenced by the absence of infectious virus in lungs, nasal passages, and brain of all animals from groups G1-G4 (Fig. 6). All unimmunized animals (groups G5-G8) developed infection, as confirmed by virus isolation from the lungs and nasal passages (Fig. 6). The mean values of virus titer in the lungs of unimmunized animals ranged from 4.57 to 7.28 log10 TCID50/mL, and in the nasal passages from 4.78 до 6.74 log10 TCID50/mL. In the brain samples from group G5, which consisted of six unimmunized animals infected with the Dubrovka strain, the infectious virus was found in all cases. In contrast, no infectious virus was detected in the brain samples of animals in other groups.
Viral RNA in the lungs, brain, heart, liver, kidneys, spleen, and blood of immunized animals (groups G1–G4) was not detected in the vast majority of animals, except for some hamsters in which RNA was detected at the limit of detection of the method. A low concentration of viral RNA was detected in the nasal passages of immunized animals, averaging 4.7 to 5.8 log10 RNA copies/mL (Fig. 7). Differences in viral RNA from nasal passages between groups of immunized and non-immunized hamsters varied depending on the strain used for challenge from 2.9 to 5.5 log10: Dubrovka strain — 3.7 (p < 0.001), Podolsk — 4.5 (p < 0.05), Otradnoe — 2.9 (p < 0.05), and FEB2 — 3.2 (p < 0.05) log10.
In the lungs of unimmunized animals (groups G5–G9), the concentration of viral RNA ranged on average from 7.60 to 9.25 log10, in the nasal passages from 8.22 to 9.34, and in the brain from 3.76 to 7.46 log10 RNA copies per mL of homogenate. In the heart, liver, kidneys, spleen, and blood of most unimmunized animals, viral RNA was also detected, but at a lower level than in the lungs and nasal passages (from 3.02 to 6.15 log10 RNA copies per mL of homogenate) (Fig. 7). Notably, when infected with the FEB2 strain, viral RNA was not detected in the liver and spleen of all animals used.
Discussion
The most outstanding achievements of public health are associated with the mass use of live vaccines: the global eradication of smallpox, the elimination of polio in most countries, a multiple reduction in the incidence of measles, rubella, mumps, rotavirus enteritis, and chickenpox. In this regard, the study of the potential of live attenuated vaccines (LAV) for the prevention of COVID-19 seems very relevant. In this study, we investigated the efficacy of the COVID-19 LAV prototype (D-D2 strain) on golden Syrian hamsters challenged with ancestral and heterologous strains of SARS-CoV-2.
Single intranasal immunization of Syrian hamsters with ca/ts/att SARS-CoV-2 D-D2 strain showed its high immunogenicity — seroconversion at 21 days p.i. was observed in all 24 immunized animals (Fig. 2, а). Antibodies titers to SARS-CoV-2 in sera of immunized hamsters as measured in ELISA, using virions of the “parental” Dubrovka strain as an immunosorbent, averaged 5 × 105. At the same time, the maximum neutralizing activity of sera was shown against the homologous Wuhan-like strain, Dubrovka strain. For the heterologous virus strains related to Delta and especially Omicron, the neutralizing activity was predictably lower or absent (Fig. 2, b), which is consistent with changes in the antigenic properties of the virus S-protein in the course of evolution and the results of previous studies [2, 3, 15, 23, 24].
Despite the reduced (or absent) neutralizing activity of post-vaccination antibodies against heterologous strains, immunization with the D-D2 strain protected hamsters against infection not only with the ancestral Dubrovka strain but also with strains belonging to Delta and Omicron VOCs. Significant protection against weight loss was shown when the animals were infected with the Dubrovka strain: immunized animals weighed 13% more (p < 0.01) than unimmunized animals at day 4 p.c. (Fig. 3). Heterologous strains, Podolsk, Otradnoe, and FEB2, showed less virulence for Syrian hamsters manifested in a slight decrease in weight of unimmunized animals. Consequently, the protection of immunized animals against weight loss of 2–4% when infected with Podolsk, Otradnoe, and FEB2 strains was not significant.
The replication level of the virus in the lungs and other organs, as well as the severity of inflammatory changes in the lungs, were more informative in assessing immunization efficacy. The absence of infectious virus in the main target organs (lung, nasal passages, and brain) in immunized animals (groups G1–G4) on the fourth day after the challenge allows us to characterize immunity to SARS-CoV-2 as "sterilizing". This conclusion is supported by the fact that in the vast majority of immunized animals, viral RNA in homogenates of lungs, brain, blood, and other organs was below the limit of detection of RT-PCR (Fig. 7). Viral RNA was detected in the nasal passages of all immunized animals, while its concentration was 2.9–4.5 log10 lower, than in unimmunized animals (Fig. 7, a). At the same time, no infectious virus was detected in the nasal passages of immunized hamsters (Fig. 6). A possible explanation for this is that the challenge was carried out by inoculation of the virus directly into the nasal passages, therefore, the nasal mucosa primarily contact with the virus and become infected. As a result, limited viral RNA replication occurs in nasal epithelial cells, but the infectious virus shedding as well as infection of lung and other organs was hindered by acquired immunity factors.
The absence of viral replication in the lungs of immunized animals is consistent with the absence of marked lung histopathology there (Figs. 4, 5). Slightly pronounced focal inflammatory changes in the bronchi revealed in the lungs of immunized hamsters infected with heterologous strains, Otradnoe and FEB2 (Figs. 4, 5), probably indicate limited virus replication due to “escape” of BA.1.1 and BA.5.2 Omicron sublineages from adaptive immunity formed after immunization with Wuhan-like strain. Thus, single intranasal immunization of Syrian hamsters with the D-D2 strain provided strong robust protection of animals against development of productive infection and pneumonia not only after homologous but also heterologous infection four weeks after immunization.
It is noteworthy that a significant viral RNA load was detected in the hearts of non-immunized animals challenged with different SARS-CoV-2 strains (up to 6.0 log10 copies of RNA/mL in group challenged with Wuhan-like virus) (Fig. 7B). These results are consistent with data from other studies showed pathological changes, viral RNA, and infectious virus in the hearts of Syrian hamsters infected with SARS-CoV-2 [25, 26]. These data are of particular importance in the context of the fact that COVID-19 increases the risk of myocarditis in humans [27, 28], which is likely due to increased expression of the ACE2 receptor in human myocytes [29].
The genetic stability of the D-D2 strain and the possibility of virulence reversion have not been investigated. Therefore, we are not yet consider the D-D2 strain as a candidate for development of COVID-19 LAV, but as a model vaccine strain to study post-vaccinal immunity.
Mass vaccination against COVID-19 has shown that licensed vaccines poorly protect against infection with the strains belonging to Omicron VOC. They provide a level of protection required against severe COVID-19 infection and death during the Omicron dominance [30, 31]. However, the post-vaccine protection against infection and symptomatic infection with the Omicron strains composed only 40–50% for mRNA vaccines (declared as the most effective vaccines) in the first 3 months p.i., and declines rapidly to 10–20% thereafter [30–33]. Booster immunization can restore this value to baseline, but the duration of the resulting immunity does not exceed 3–6 months [31, 32, 34–36]. Licensed vaccines (inactivated, vector, recombinant, and mRNA vaccines) based on the S-protein of SARS-CoV-2 stimulate both humoral and cellular immunity ([37–39]; however, apparently their efficacy is mainly determined by the induction of neutralizing antibodies. Variability of the S-protein and the emergence of new VOCs leads to a rapid escape of the virus from immunological surveillance and a decrease in the effectiveness of such vaccines [2, 3, 23, 24, 40]. The ongoing evolution of the Omicron VOC has led to the emergence of such of its sublineages that escape adaptive immunity induced even by previous infection with parental Omicron sublineages. Thus, sublineages BF.7, BQ.1, and XBB (which appeared after sublineage BA.4/5), escape neutralization by antibodies, antibody-dependent cellular cytotoxicity, and phagocytosis, induced by breakthrough infection with sublineage BA.1 [41]. In addition, the sublineages BF.7 and BQ.1 have the greatest resistance to neutralization by a panel of 77 monoclonal antibodies that effectively neutralize the Wuhan-like virus [42].
The higher efficacy of intranasally administered LAV is based on the same mechanisms that are involved in the development of adaptive immunity during natural respiratory virus infection [37, 43, 44]. Combined activation of the humoral and cellular components of the systemic and mucosal (local) immune defenses can provide effective protection against SARS-CoV-2 infection [8, 37, 45]. Furthermore, an immune response is developed against all viral proteins of LAV, both structural and nonstructural, increasing the efficacy. Thus, structural proteins M and N are highly immunogenic and, together with nonstructural proteins, are more conserved than the S-protein. Furthermore, many T-cell epitopes of phylogenetically related species of coronaviruses and various SARS-CoV-2 variants are located not only in the S-protein [46, 47]. Thus, J. Zhao et al., using peripheral blood mononuclear cells isolated from COVID-19 patients, identified 5 immunodominant T-cell epitopes in the N-protein of SARS-CoV-2 [47]. It should also be noted that protective immunity mediated by T-cells is less dependent on mutations that determine the formation of new SARS-CoV-2 VOCs [48, 49]. Indeed the appearance of new SARS-CoV-2 VOCs is determined mainly by mutations in the most variable S-protein, whereas T-cell epitopes are present not only in the S-protein, but also in more conserved viral proteins.
In recent studies, the immunogenicity and efficacy of attenuated SARS-CoV-2 strains were investigated on animal models of coronavirus infection, such as golden Syrian hamsters (Mesocricetus auratus), Phodopus roborovskii hamsters and K18-hACE2 transgenic mice line. Immunization of susceptible animals with attenuated strains of SARS-CoV-2 has been found to provide highly effective protection against homologous infection and development of pneumonia [9–17]. However, the potential for cross-protective activity of LAVs against heterologous variants of the virus has not been extensively investigated. J. Trimpert et al. showed that immunization with attenuated SARS-CoV-2 leads to the development of immunity in laboratory animals when infected not only with the parental strain of the virus but also with unrelated Alpha and Beta VOC strains [50]. A. Yoshida et al. showed the development of cross-protection against infection with the Omicron (BA.1) VOC strain in hamsters immunized with the recombinant attenuated virus with the S-protein gene "transferred" from Omicron [13]. Our study shows that the attenuated Wuhan-like SARS-CoV-2 strain can provide highly effective protection not only against the homologous challenge but also when challenged with heterologous strains belonging to Delta VOC, BA.1.1 and BA.5.2 Omicron VOC sublineages.
The cross-protection shown in our study was predictable because at the time the study was designed it was known that naturally acquired SARS-CoV-2 infection prevents up to 90% of reinfection with Alpha, Beta, or Delta VOCs and 56% of reinfection with Omicron VOC strains, while most reinfection cases occur only one year after the primary disease [51]. Furthermore, the protective efficacy of primary infection against the development of severe disease or death in reinfection with the Omicron is 97.3% (95% CI 94.9–98.6%), regardless of the virus variant that caused the primary infection [52]. The extremely low rate of severe and fatal cases of reinfection potentially points out that COVID-19 LAV can provide effective protection against pneumonia and death caused by heterologous strains. The high potential of LAVs for COVID-19 prevention is supported by the observation that hybrid immunity (vaccination followed by a breakthrough infection) and SARS-CoV-2 reinfections reduce the risk of subsequent infections caused by the Omicron strains by 60% and 85%, respectively [53]. Meanwhile, boosterization by breakthrough Omicron infection induces higher levels of memory B-cell and SARS-CoV-2-specific T-cells, particularly against the Omicron strains, compared with booster immunization with inactivated or vector vaccines [54].
Noteworthy, practically all developers of LAVs against COVID-19 achieve effective protection against challenges with the virulent strain by intranasal administration [9–17]. In this regard, the successful mass use of mucosal vaccines such as live polio and rotavirus vaccines (administered orally) and live influenza vaccines (administered intranasally) are revealing. Oral and intranasal administration of these vaccines provides not only induction of systemic cellular and humoral adaptive immune response but also the formation of mucosal (local) immunity, including secretion of specific IgA-antibodies in the respiratory or intestinal mucosa. During intranasal immunization followed by infection with a virulent strain, specific secretory IgA-antibodies neutralize the virus directly on the mucosa of the respiratory tract, which is the site of entry of infection, suppressing its adhesive ability and reducing the efficiency of transmission [8, 55, 56].
Conclusions
The results of the present study showed that single intranasal immunization of Syrian hamsters with attenuated Wuhan-like strain of SARS-CoV-2 provides strong robust protection of animals against infection and development of pneumonia when challenged not only with homologous virus but also with heterologous strains belonging to Delta (AY.122) and Omicron (sublineages BA.1 and BA.5.2) VOCs. Thus, creation of attenuated SARS-CoV-2 strains is a promising strategy for the development of highly effective live vaccines against COVID-19 on their basis.
1 URL: https://www.virosin.org/tcid50/TCID50.html
About the authors
Evgeny B. Faizuloev
I.I. Mechnikov Research Institute of Vaccines and Sera; Russian Medical Academy of Continuous Professional Education
Email: faizuloev@mail.ru
ORCID iD: 0000-0001-7385-5083
Cand. Sci. (Biol.), Head, Applied virology laboratory, I. Mechnikov Research Institute of Vaccines and Sera, Moscow, Russia; senior lecturer, Department of virology, Russian Medical Academy of Continuous Professional Education
Russian Federation, Moscow; MoscowAnastasiia V. Gracheva
I.I. Mechnikov Research Institute of Vaccines and Sera
Email: faizuloev@mail.ru
ORCID iD: 0000-0001-8428-4482
researcher, Applied virology laboratory, I. Mechnikov Research Institute of Vaccines and Sera
Russian Federation, MoscowEkaterina R. Korchevaya
I.I. Mechnikov Research Institute of Vaccines and Sera
Email: faizuloev@mail.ru
ORCID iD: 0000-0002-6417-3301
junior researcher, Applied virology laboratory, I. Mechnikov Research Institute of Vaccines and Sera
Russian Federation, MoscowYulia I. Ammour
I.I. Mechnikov Research Institute of Vaccines and Sera
Email: faizuloev@mail.ru
ORCID iD: 0000-0003-0223-5738
Cand. Sci. (Biol.), Head, Oncolytic viruses laboratory, I. Mechnikov Research Institute of Vaccines and Sera
Russian Federation, MoscowDaria I. Smirnova
I.I. Mechnikov Research Institute of Vaccines and Sera
Email: faizuloev@mail.ru
ORCID iD: 0000-0001-7325-0834
junior researcher, Applied virology laboratory, I. Mechnikov Research Institute of Vaccines and Sera
Russian Federation, MoscowDarya M. Khokhlova
I.I. Mechnikov Research Institute of Vaccines and Sera
Email: faizuloev@mail.ru
ORCID iD: 0009-0003-5745-7589
junior researcher, Applied virology laboratory, I. Mechnikov Research Institute of Vaccines and Sera
Russian Federation, MoscowAndrey O. Drokov
I.M. Sechenov First Moscow State Medical University (Sechenov University)
Email: faizuloev@mail.ru
ORCID iD: 0009-0006-3300-8078
intern, Applied virology laboratory, I. Mechnikov Research Institute of Vaccines and Sera
Russian Federation, MoscowAndrey A. Pankratov
P.A. Hertsen Moscow Oncology Research Institute
Email: faizuloev@mail.ru
ORCID iD: 0000-0001-7291-9743
Cand. Sci. (Biol.), Head, Department of experimental pharmacology and toxicology, P.A. Hertsen Moscow Oncology Research Institute
Russian Federation, MoscowGalina V. Trunova
P.A. Hertsen Moscow Oncology Research Institute
Author for correspondence.
Email: faizuloev@mail.ru
ORCID iD: 0000-0003-2917-4496
Cand. Sci. (Biol.), senior researcher, Department of experimental pharmacology and toxicology, P.A. Hertsen Moscow Oncology Research Institute
Russian Federation, MoscowVarvara A. Khokhlova
P.A. Hertsen Moscow Oncology Research Institute
Email: faizuloev@mail.ru
ORCID iD: 0000-0002-0339-2068
junior researcher, Department of experimental pharmacology and toxicology, P.A. Hertsen Moscow Oncology Research Institute
Russian Federation, MoscowMaria S. Vorontsova
P.A. Hertsen Moscow Oncology Research Institute
Email: faizuloev@mail.ru
ORCID iD: 0000-0002-9320-1746
Cand. Sci. (Biol.), junior researcher, Department of experimental pharmacology and toxicology, P.A. Hertsen Moscow Oncology Research Institute
Russian Federation, MoscowIrina A. Leneva
I.I. Mechnikov Research Institute of Vaccines and Sera
Email: faizuloev@mail.ru
ORCID iD: 0000-0002-7755-2714
D. Sci. (Biol.), Head, Experimental virology laboratory, I. Mechnikov Research Institute of Vaccines and Sera
Russian Federation, MoscowOksana A. Svitich
I.I. Mechnikov Research Institute of Vaccines and Sera; I.M. Sechenov First Moscow State Medical University (Sechenov University)
Email: faizuloev@mail.ru
ORCID iD: 0000-0003-1757-8389
D. Sci. (Med.), Prof., Corresponding Member of RAS, Director, I. Mechnikov Research Institute of Vaccines and Sera, Moscow, Russia; Professor, Department of microbiology, virology and immunology, I.M. Sechenov First Moscow State Medical University (Sechenov University)
Russian Federation, Moscow; MoscowVitaly V. Zverev
I.I. Mechnikov Research Institute of Vaccines and Sera; I.M. Sechenov First Moscow State Medical University (Sechenov University)
Email: faizuloev@mail.ru
ORCID iD: 0000-0001-5808-2246
D. Sci. (Biol.), Prof., Academician of RAS, Scientific director, I. Mechnikov Research Institute of Vaccines and Sera, Moscow, Russia; Head, Department of microbiology, virology and immunology, I.M. Sechenov First Moscow State Medical University (Sechenov University)
Russian Federation, Moscow; MoscowReferences
- Feikin D.R., Higdon M.M., Abu-Raddad L.J., et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression. Lancet. 2022;399(10328):924–44. DOI: https://doi.org/10.1016/s0140-6736(22)00152-0
- Bowen J.E., Addetia A., Dang H.V., et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science. 2022;377(6608):890–4. DOI: https://doi.org/10.1126/science.abq0203
- Dejnirattisai W., Huo J., Zhou D., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467–84.e15. DOI: https://doi.org/10.1016/j.cell.2021.12.046
- Xiang T., Wang J., Zheng X. The humoral and cellular immune evasion of SARS-CoV-2 Omicron and sub-lineages. Virol. Sin. 2022;37(6):786–95. DOI: https://doi.org/10.1016/j.virs.2022.11.007
- Chalkias S., Harper C., Vrbicky K., et al. A bivalent omicron-containing booster vaccine against COVID-19. N. Engl. J. Med. 2022;387(14):1279–91. DOI: https://doi.org/10.1056/NEJMoa2208343
- Chen J.M. Should the world collaborate imminently to develop neglected live-attenuated vaccines for COVID-19? J. Med. Virol. 2022;94(1):82–7. DOI: https://doi.org/10.1002/jmv.27335
- Goławski M., Lewandowski P., Jabłońska I., Delijewski M. The reassessed potential of SARS-CoV-2 attenuation for COVID-19 vaccine development – a systematic review. Viruses. 2022;14(5):991. DOI: https://doi.org/10.3390/v14050991
- Nouailles G., Adler J.M., Pennitz P., et al. Live-attenuated vaccine sCPD9 elicits superior mucosal and systemic immunity to SARS-CoV-2 variants in hamsters. Nat. Microbiol. 2023;8(5):860–74. DOI: https://doi.org/10.1038/s41564-023-01352-8
- Liu S., Stauft C.B., Selvaraj P., et al. Intranasal delivery of a rationally attenuated SARS-CoV-2 is immunogenic and protective in Syrian hamsters. Nat. Commun. 2022;13(1):6792. DOI: https://doi.org/10.1038/s41467-022-34571-4
- Liu Y., Zhang X., Liu J., et al. A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions. Nat. Commun. 2022;13(1):4337. DOI: https://doi.org/10.1038/s41467-022-31930-z
- Trimpert J., Dietert K., Firsching T.C., et al. Development of safe and highly protective live-attenuated SARS-CoV-2 vaccine candidates by genome recoding. Cell Rep. 2021;36(5):109493. DOI: https://doi.org/10.1016/j.celrep.2021.109493
- Ye Z.W., Ong C.P., Tang K., et al. Intranasal administration of a single dose of a candidate live attenuated vaccine derived from an NSP16-deficient SARS-CoV-2 strain confers sterilizing immunity in animals. Cell. Mol. Immunol. 2022;19(5):588–601. DOI: https://doi.org/10.1038/s41423-022-00855-4
- Yoshida A., Okamura S., Torii S., et al. Versatile live-attenuated SARS-CoV-2 vaccine platform applicable to variants induces protective immunity. iScience. 2022;25(11):105412. DOI: https://doi.org/10.1016/j.isci.2022.105412
- Abdoli M., Shafaati M., Ghamsari L.K., Abdoli A. Intranasal administration of cold-adapted live-attenuated SARS-CoV-2 candidate vaccine confers protection against SARS-CoV-2. Virus Res. 2022;319:198857. DOI: https://doi.org/10.1016/j.virusres.2022.198857
- Faizuloev E., Gracheva A., Korchevaya E., et al. Cold-adapted SARS-CoV-2 variants with different temperature sensitivity exhibit an attenuated phenotype and confer protective immunity. Vaccine. 2023;41(4):892–902. DOI: https://doi.org/10.1016/j.vaccine.2022.12.019
- Seo S.H., Jang Y. Cold-adapted live attenuated SARS-CoV-2 vaccine completely protects human ACE2 transgenic mice from SARS-CoV-2 infection. Vaccines (Basel). 2020;8(4):584. DOI: https://doi.org/10.3390/vaccines8040584
- Xu J., Liu M., Niu X., et al. The cold-adapted, temperature-sensitive SARS-CoV-2 strain TS11 is attenuated in Syrian hamsters and a candidate attenuated vaccine. Viruses. 2022;15(1): 95. DOI: https://doi.org/10.3390/v15010095
- Файзулоев Е.Б., Корчевая Е.Р., Грачева А.В. и др. Биологическая характеристика холодоадаптированных вариантов коронавируса SARS-CoV-2. Журнал микробиологии, эпидемиологии и иммунобиологии. 2022;99(4):397–409. Faizuloev E.B., Korchevaya E.R., Gracheva A.V., et al. Biological characterization of cold-adapted SARS-CoV-2 variants. Journal of Microbiology, Epidemiology and Immunobiology. 2022;99(4):397–409. DOI: https://doi.org/10.36233/0372-9311-280 EDN: https://elibrary.ru/llgegh
- Грачёва А.В., Корчевая Е.Р., Кудряшова А.М. и др. Адаптация МТТ-теста для определения нейтрализующих антител к вирусу SARS-CoV-2. Журнал микробиологии, эпидемиологии и иммунобиологии. 2021;98(3):253–65. Gracheva A.V., Korchevaya E.R., Kudryashova A.M., et al. Adaptation of the MTT assay for detection of neutralizing antibodies against the SARS-CoV-2 virus. Journal of Microbiology, Epidemiology and Immunobiology. 2021;98(3):253–65. DOI: https://doi.org/10.36233/0372-9311-136 EDN: https://elibrary.ru/jglovv
- Gracheva A.V., Korchevaya E.R., Ammour Y.I., et al. Immunogenic properties of SARS-CoV-2 inactivated by ultraviolet light. Arch. Virol. 2022;167(11):2181–91. DOI: https://doi.org/10.1007/s00705-022-05530-7
- Chan J.F.W., Yip C.C.Y., To K.K.W., et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020;58(5):e00310-20. DOI: https://doi.org/10.1128/JCM.00310-20
- Gruber A.D., Osterrieder N., Bertzbach L.D., et al. Standardization of reporting criteria for lung pathology in SARS-CoV-2-infected hamsters: what matters? Am. J. Respir. Cell Mol. Biol. 2020;63(6):856–9. DOI: https://doi.org/10.1165/rcmb.2020-0280LE
- Aiano F., Ireland G., Baawuah F., et al. Antibody persistence after primary SARS-CoV-2 infection and protection against future variants including omicron in adolescents: national, prospective cohort study. Pediatr. Infect. Dis. J. 2023;42(6):496–502. DOI: https://doi.org/10.1097/inf.0000000000003890
- Wang Y., Ma Y., Xu Y., et al. Resistance of SARS-CoV-2 Omicron variant to convalescent and CoronaVac vaccine plasma. Emerg. Microbes Infect. 2022;11(1):424–7. DOI: https://doi.org/10.1080/22221751.2022.2027219
- Daems M., Liesenborghs L., Boudewijns R., et al. SARS-CoV-2 infection causes prolonged cardiomyocyte swelling and inhibition of HIF1α translocation in an animal model COVID-19. Front. Cardiovasc. Med. 2022;9:964512. DOI: https://doi.org/10.3389/fcvm.2022.964512
- Jones E.A.V. Mechanism of COVID-19-induced cardiac damage from patient, in vitro and animal studies. Curr. Heart Fail. Rep. 2023;20(5):451–60. DOI: https://doi.org/10.1007/s11897-023-00618-w
- Ishisaka Y., Watanabe A., Aikawa T., et al. Overview of SARS-CoV-2 infection and vaccine associated myocarditis compared to non-COVID-19-associated myocarditis: A systematic review and meta-analysis. Int. J. Cardiol. 2024;395:131401. DOI: https://doi.org/10.1016/j.ijcard.2023.131401
- Thaker R., Faraci J., Derti S., Schiavone J.F. Myocarditis in SARS-CoV-2: a meta-analysis. Cureus. 2023;15(10):e48059. DOI: https://doi.org/10.7759/cureus.48059
- Liu H., Gai S., Wang X., et al. Single-cell analysis of SARS-CoV-2 receptor ACE2 and spike protein priming expression of proteases in the human heart. Cardiovasc. Res. 2020;116(10):1733–41. DOI: https://doi.org/10.1093/cvr/cvaa191
- Menegale F., Manica M., Zardini A., et al. Evaluation of waning of SARS-CoV-2 vaccine-induced immunity: a systematic review and meta-analysis. JAMA Netw. Open. 2023;6(5):e2310650. DOI: https://doi.org/10.1001/jamanetworkopen.2023.10650
- Paul P., El-Naas A., Hamad O., et al. Effectiveness of the pre-Omicron COVID-19 vaccines against Omicron in reducing infection, hospitalization, severity, and mortality compared to Delta and other variants: A systematic review. Hum. Vaccin. Immunother. 2023;19(1):2167410. DOI: https://doi.org/10.1080/21645515.2023.2167410
- Amir O., Goldberg Y., Mandel M., et al. Protection against Omicron BA.1/BA.2 severe disease 0–7 months after BNT162b2 booster. Commun. Biol. 2023;6(1):315. DOI: https://doi.org/10.1038/s42003-023-04669-6
- Lau J.J., Cheng S.M.S., Leung K., et al. Real-world COVID-19 vaccine effectiveness against the Omicron BA.2 variant in a SARS-CoV-2 infection-naive population. Nat. Med. 2023;29(2):348–57. DOI: https://doi.org/10.1038/s41591-023-02219-5
- Chemaitelly H., Ayoub H.H., AlMukdad S., et al. Duration of mRNA vaccine protection against SARS-CoV-2 Omicron BA.1 and BA.2 subvariants in Qatar. Nat. Commun. 2022;13(1):3082. DOI: https://doi.org/10.1038/s41467-022-30895-3
- Espíndola O.M., Fuller T.L., de Araújo M.F., et al. Reduced ability to neutralize the Omicron variant among adults after infection and complete vaccination with BNT162b2, ChAdOx1, or CoronaVac and heterologous boosting. Sci. Rep. 2023;13(1):7437. DOI: https://doi.org/10.1038/s41598-023-34035-9
- Huiberts A.J., de Gier B., Hoeve C.E., et al. Vaccine effectiveness of primary and booster COVID-19 vaccinations against SARS-CoV-2 infection in the Netherlands from July 12, 2021 to June 6, 2022: A prospective cohort study. Int. J. Infect. Dis. 2023;133:36–42. DOI: https://doi.org/10.1016/j.ijid.2023.04.401
- Корчевая Е.Р., Грачева А.В., Дьяков И.Н. и др. Живые аттенуированные вакцины против COVID-19: подходы к разработке и перспективы клинического применения. Журнал микробиологии, эпидемиологии и иммунобиологии. 2023;100(3):225–36. Korchevaya E.R., Gracheva A.V., Dyakov I.N., et al. Live attenuated COVID-19 vaccines: approaches to development and prospects for clinical use. Journal of Microbiology, Epidemiology and Immunobiology. 2023;100(3):225–36. DOI: https://doi.org/10.36233/0372-9311-404 EDN: https://elibrary.ru/psdxzr
- Park H., Park M.S., Seok J.H., et al. Insights into the immune responses of SARS-CoV-2 in relation to COVID-19 vaccines. J. Microbiol. 2022;60(3):308–20. DOI: https://doi.org/10.1007/s12275-022-1598-x
- Rahman M.M., Masum M.H.U., Wajed S., Talukder A. A comprehensive review on COVID-19 vaccines: development, effectiveness, adverse effects, distribution and challenges. Virusdisease. 2022;33(1):1–22. DOI: https://doi.org/10.1007/s13337-022-00755-1
- Seidel A., Jacobsen E.M., Fabricius D., et al. Serum neutralizing capacity and T-cell response against the omicron BA.1 variant in seropositive children and their parents one year after SARS-CoV-2 infection. Front. Pediatr. 2023;11:1020865. DOI: https://doi.org/10.3389/fped.2023.1020865
- Guo L., Zhang Q., Zhong J., et al. Omicron BA.1 breakthrough infections in inactivated COVID-19 vaccine recipients induced distinct pattern of antibody and T cell responses to different Omicron sublineages. Emerg. Microbes Infect. 2023;12(1):2202263. DOI: https://doi.org/10.1080/22221751.2023.2202263
- Chen J., Yang J., Chang F., et al. Identification of broad neutralizing antibodies against Omicron subvariants from COVID-19 convalescents and vaccine recipients. Virol. Sin. 2023;38(2): 313–6. DOI: https://doi.org/10.1016/j.virs.2023.01.005
- Brown B., Ojha V., Fricke I., et al. Innate and adaptive immunity during SARS-CoV-2 infection: biomolecular cellular markers and mechanisms. Vaccines (Basel). 2023;11(2):408. DOI: https://doi.org/10.3390/vaccines11020408
- Shen J., Fan J., Zhao Y., et al. Innate and adaptive immunity to SARS-CoV-2 and predisposing factors. Front. Immunol. 2023; 14:1159326. DOI: https://doi.org/10.3389/fimmu.2023.1159326
- Miteva D., Peshevska-Sekulovska M., Snegarova V., et al. Mucosal COVID-19 vaccines: Risks, benefits and control of the pandemic. World J. Virol. 2022;11(5):221–36. DOI: https://doi.org/10.5501/wjv.v11.i5.221
- Mateus J., Grifoni A., Tarke A., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370(6512):89–94. DOI: https://doi.org/10.1126/science.abd3871
- Zhao J., Wang L., Schank M., et al. SARS-CoV-2 specific memory T cell epitopes identified in COVID-19-recovered subjects. Virus Res. 2021;304:198508. DOI: https://doi.org/10.1016/j.virusres.2021.198508
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–80. DOI: https://doi.org/10.1016/j.cell.2021.01.007
- Tarke A., Sidney J., Kidd C.K., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2(2):100204. DOI: https://doi.org/10.1016/j.xcrm.2021.100204
- Trimpert J., Adler J.M., Eschke K., et al. Live attenuated virus vaccine protects against SARS-CoV-2 variants of concern B.1.1.7 (Alpha) and B.1.351 (Beta). Sci. Adv. 2021;7(49):eabk0172. DOI: https://doi.org/10.1126/sciadv.abk0172
- Altarawneh H.N., Chemaitelly H., Hasan M.R., et al. Protection against the Omicron variant from previous SARS-CoV-2 infection. N. Engl. J. Med. 2022;386(13):1288–90. DOI: https://doi.org/10.1056/nejmc2200133
- Chemaitelly H., Nagelkerke N., Ayoub H.H., et al. Duration of immune protection of SARS-CoV-2 natural infection against reinfection. J. Travel Med. 2022;29(8):taac109. DOI: https://doi.org/10.1093/jtm/taac109
- Sun K., Tempia S., Kleynhans J., et al. Rapidly shifting immunologic landscape and severity of SARS-CoV-2 in the Omicron era in South Africa. Nat Commun. 2023;14(1):246. doi: 10.1038/s41467-022-35652-0
- Yu P., Liu Z., Zhu Z., et al. Omicron variants breakthrough infection elicited higher specific memory immunity than third dose booster in healthy vaccinees. Virol. Sin. 2023;38(2):233–43. DOI: https://doi.org/10.1016/j.virs.2022.12.008
- Jacobson R.M., Poland G.A. Universal vaccination of healthy children against influenza: a role for the cold-adapted intranasal influenza vaccine. Pediatr. Drugs. 2002;4(1):65–71. DOI: https://doi.org/10.2165/00128072-200204010-00007
- Nian X., Zhang J., Huang S., et al. Development of nasal vaccines and the associated challenges. Pharmaceutics. 2022;14(10):1983. DOI: https://doi.org/10.3390/pharmaceutics14101983
Supplementary files