Subgingival microbiome in periodontal disease and comorbid pathology (meta-analysis)
- Authors: Tsareva T.V.1, Balmasova I.P.1, Tsarev V.N.1
-
Affiliations:
- Russian University of Medicine
- Issue: Vol 101, No 2 (2024)
- Pages: 281-292
- Section: REVIEWS
- URL: https://microbiol.crie.ru/jour/article/view/18538
- DOI: https://doi.org/10.36233/0372-9311-500
- EDN: https://elibrary.ru/mwauhy
- ID: 18538
Cite item
Abstract
The problem of chronic periodontitis (CP) is actively discussed due to the recognition of the fact that periodontal microbial damage is closely related to a number of systemic diseases and probably plays an important role in the occurrence of comorbid pathology.
The aim of the meta-analysis was to characterize the composition of the subgingival microbiome and to determine the peculiarities of the formation of associations of the new periodontal pathogen Filifactor alocis with other I and II order periodontal pathogenic bacteria, as well as with the commensal bacteria colonizing this biotope.
The study presents data of patient examination with obligatory use of polymerase chain reaction methods and sequencing of 16S rRNA genes in 1529 healthy individuals and 2394 patients with CP, 136 patients with CP and concomitant atherosclerosis, 258 patients with CP and concomitant type 2 diabetes mellitus. It was confirmed that the basis of the oral microbiome under normal circumstances is composed of representatives of microaerophilic streptococci, corynebacteria, lactobacilli, as well as representatives of the Veillonella and Sphingobacterium genera. 16S sequencing and bioinformatic analysis allowed us to specify the taxonomic place of the new pathogen F. alocis, as well as representatives of normal microbiota in CP and comorbid somatic pathology.
Full Text
Introduction
Chronic generalized periodontitis (CP) is a severe lesion of the gums and underlying periodontal tissues with a recurrent course, which is the main cause of tooth loss in middle and old aged people. In terms of prevalence, this pathology is the leading dental problem in the world among able-bodied adults, although significant variations in frequency have been found depending on age, region, climatic and social conditions (frequency varies from 60 to 90%) [1–3]. The socio-medical significance of the problem of CP is constantly increasing, especially in connection with the growth of the disease of aggressive periodontitis developing at a young age [4, 5]. The problem of CP is actively discussed at various congresses held under the auspices of WHO due to the recognition of the fact that microbial damage of the periodontium is closely related to a number of systemic diseases and likely plays an important role in the occurrence of comorbid pathology [5–7].
The ability of microorganisms to colonize the oral mucosa, periodontal tissues and tooth surfaces with the formation of a multicomponent structured microbial biofilm is extremely important from a biological point of view. Bacteria perform numerous metabolic, barrier and protective functions, contribute to the integrity of periodontal tissues, increase the proliferation of epithelial cells, prepare the mucosal barrier to potential pathogen infiltration through the production of antimicrobial peptides and cytokines [6].
The key moment in the development of infectious-inflammatory lesions of periodontal tissues is the colonization of microbial biofilm by priority pathogens capable of intracellular parasitism and called periodontal pathogenic. The localization of this biofilm is in the area of the dento-gingival sulcus, and as the destructive process progresses, it is localized in the forming periodontal pocket [8, 9].
Thus, the topic at hand is the subgingival microbiome formed when the biofilm is colonized by both normal biotic and periodontal pathogenic bacterial species.
In 1998, S.S. Socransky et al. proposed a classification based on the study of the degree of frequency of identification of microflora representatives in the focus of inflammation: red, orange and other complexes, as well as to a greater extent representing normal and transient microflora — yellow, green and violet were identified. Porphyromonas gingivalis, Tannerlla forsythia (according to the old nomenclature — Bacteroides forsytus), Treponema denticola [10], in particular, are referred to the red complex. However, at present, this classification has more historical significance, since some pathogens of aggressive periodontitis, for example, toxigenic serotype b actinobacillus Aggregatibacter actinomycetemcomitans, were described somewhat later [11].
After the I Congress of Periodontologists of Russia in 2005, the concept of periodontal pathogenic bacteria of the first order was first introduced in the Russian literature, which possess a combination of three leading features: vertical and horizontal transmission of the infectious agent from person to person, the ability to intracellular parasitism, and toxicogenicity (primarily the production of exotoxins), in contrast to periodontal pathogens of the second order, which do not have a full set of these features. Three pathogens, P. gingivalis, T. forsythia, and A. actinomycetemcomitans, toxigenic serotype b, were classified as periodontal pathogenic species of the I order [8, 9, 12]. In further studies, diagnostically significant microbial number indices for different species of periodontal pathogenic bacteria of I and II orders were established, which with certain adjustments are presented in Table 1.
Table 1. Quantitative parameters of the content of periodontal pathogenic bacteria of I and II orders in the gingival biofilm under normal circumstances [6, 13]
Periodontal pathogen group | Group traits | Periodontal pathogen type | Quantity, CFU/ml |
Periodontal pathogenic species of the I order | Representatives of these species are pathogens of periodontitis; normally absent in a healthy person (except for healthy carriage in 6–12% of cases) | A. actinomycetemcomitans | > 102 |
B. (Tannerella) forsythia | > 102 | ||
P. gingivalis | > 102 | ||
Periodontal pathogenic species of the II order | Representatives of these species on the gums are normally absent or determined in small numbers. Exceeding the specified number is a sign of periodontitis development | Prevotella intermedia | > 102 |
Treponema denticola | > 103 | ||
Fusobacterium nucleatum | > 103 | ||
Filifactor alocis | > 103 | ||
Actinomyces israelii | > 104 | ||
Parvimonas micros | > 104 | ||
Streptococcus intermedius | > 104 | ||
Eykenella corrodens | > 103 | ||
Selenomonas spp. | > 103 | ||
Wolinella recta | > 103 | ||
Candida albicans, C. krusei, C. glabrata | > 102 |
In the development of periodontal diseases, synergistic and/or mutualistic interactions between oral pathogens (periodontal pathogens) are formed quite often, which are believed to contribute to the onset and progression of the disease. However, it should not be forgotten that such relationships are based on the potentiation of the action of their virulence factors as agents of the infectious process [6, 13]. While the etiologic role of P. gingivalis as a key periodontal pathogen is beyond doubt [14], the significance of some other hard-to-cultivate bacteria, which have begun to be studied according to modern molecular research methods, remains up to debate [15-17].
One of these microbes appears to be F. alocis, which has been relatively recently excluded from the Fusobacteria class, type Firmicutes, into a separate genus represented by a single species. As emphasized by E. Aja et al., F. alocis is a new member of the periodontal microbiome, and it is currently proposed to be used as a diagnostic indicator of periodontal diseases [18]. However, due to the lack of genetic tools to study this microorganism, until recently little is known about its characteristics and virulence, and its importance in comorbid pathology has been described in very few publications [19]. This circumstance prompted us, in recent studies, to create a diagnostic set of primers and probes for multiplex polymerase chain reaction (PCR) for the detection of this species of periodontal pathogenic bacteria [20, 21]. On the other hand, there is a clear need to analyze the available data on the composition and relationships of the components of the subgingival microbiome.
The aim of the meta-analysis was to characterize the composition of the subgingival microbiome and to determine the features of the formation of associations of the new periodontal pathogen F. alocis with other I and II order periodontal pathogenic bacteria, as well as with the commensal bacteria colonizing this biotope.
Materials and methods
Searches were conducted in MEDLINE (via PubMed), EMBASE, and Cochrane Library databases from 2005 to October 2023 using keywords identified in the study objectives using a search strategy that met the developed inclusion and exclusion criteria.
Inclusion criteria:
1) microbiome studies using bacterial 16S rRNA gene sequencing, including meta-analyses and systematic reviews that compared results obtained by microbiologic and molecular biologic methods (percentages of occurrence of bacterial types, genera, species) in subgingival plaque samples obtained:
- from systemically healthy patients without periodontal disease;
- from patients with CP without systemic pathology;
- from patients with CP with concomitant verified atherosclerosis (AT);
- from patients with CP with concomitant verified type 2 diabetes mellitus (T2D);
2) only studies indicating the detection of a relatively new difficult-to-cultivate periodontal pathogenic species, F. alocis, in the microbiome were taken into account.
Exclusion criteria:
- studies published in languages other than Russian or English;
- absence of primary data;
- absence of data on samples from the gingival sulcus of systemically healthy people in the groups of healthy people and/or from periodontal pockets with a depth of 5 mm or more in the groups of CP patients, including those with concomitant pathology;
- studies that evaluated only patients with gingivitis without signs of periodontitis, with aggressive or refractory periodontitis.
The search found a total of 1437 studies. After title screening, 1211 papers were excluded and 126 were screened. After reading the abstracts, 12 studies were excluded and 114 full-text publications were comprehensively evaluated. After reading these studies, 92 were excluded for not meeting the inclusion criteria. Thus, results from 22 literature sources were included in this study.
Results
Once the appropriate array was obtained, data extraction was performed with the following additional information labeled:
- study location;
- characteristics of the participants (in terms of clinical periodontal condition and presence of comorbid pathology);
- type of microbiologic evaluation (study method);
- microbiologic results (in percentages) for individual bacterial taxa in different study groups.
Bacterial types, genera, species were subsequently indexed according to the taxonomic index of the US National Center for Biotechnology Information1.
The methodological features, year and place of research in the literature sources used for the meta-analysis are summarized and presented in Table 2.
Table 2. Information about the characteristics of the studies included in the meta-analysis
Study type | Country | Study group | n | Source |
Original | USA | Healthy patients | 15 | [22] |
Patients with CP | 15 | |||
Original | USA | Healthy patients | 29 | [23] |
Patients with CP | 29 | |||
Original | USA | Healthy patients | 5 | [24] |
Patients with CP | 2 | |||
Original | USA, Chile | Healthy patients | 10 | [2] |
Patients with CP | 22 | |||
Systemic review | USA | Healthy patients | 98 | [25] |
Patients with CP | 24 | |||
Original | Brazil, Canada | Patients with CP and AT | 18 | [26] |
Original | Brazil | Healthy patients | 27 | [27] |
Patients with CP | 59 | |||
Original | Brazil | Healthy patients | 912 | [28] |
Patients with CP | 1918 | |||
Original | Korea | Healthy patients | 12 | [29] |
Patients with CP | 10 | |||
Original | Russia | Patients with CP and AT | 28 | [30] |
Original | USA | Healthy patients | 76 | [31] |
Patients with CP | 76 | |||
Original | USA, India | Healthy patients | 75 | [32] |
Patients with CP | 50 | |||
Patients with CP and T2D | 50 | |||
Original | USA | Healthy patients | 97 | [33] |
Patients with CP and T2D | 98 | |||
Original | Brazil, USA | Healthy patients | 98 | [34] |
Original | Mexico | Healthy patients | 59 | [35] |
Patients with CP | 67 | |||
Patients with CP and T2D | 38 | |||
Original | Saudi Arabia | Healthy patients | 19 | [36] |
Patients with CP and T2D | 15 | |||
Original | South Africa | Healthy patients | 32 | [37] |
Patients with CP | 32 | |||
Patients with CP and T2D | 32 | |||
Original | Russia | Healthy patients | 16 | [21] |
Patients with CP | 15 | |||
Patients with CP and T2D | 15 | |||
Original | Tunisia | Healthy patients | 10 | [38] |
Patients with CP and AT | 20 | |||
Original | Italy | Healthy patients | 15 | [39] |
Patients with CP and AT | 15 | |||
Original | India | Patients with CP and AT | 12 | [40] |
Original | Japan | Patients with CP and AT | 43 | [41] |
Note. 16S rRNA genes were sequenced in all studies.
Among the literature sources included in the study, there were 2 systematic reviews showing numerical data pooled from several sources, and the remaining sources contained the results of original studies. Quantitative data used in the meta-analysis were contained either as part of the graphical material or as supplementary information to the publication.
The generalized characteristics of the contingents of individuals analyzed from the original studies are presented in Table 3. The number of examined healthy individuals and patients suffering from CP was contained in 16 and 12 sources, respectively, and was quite significant (1529 and 2394 individuals). As for the groups of patients with CP associated with systemic pathological processes, only 6 sources containing data on the composition of the subgingival microbiome in 136 people with the association of CP and AT, and 6 sources based on the results of similar studies analyzing the data of 258 people with the association of CP and T2D met the search conditions.
Table 3. Cumulative characteristics of the study population included in the meta-analysis
Analyzed attributes | Study groups | ||||
healthy patients | patients with CP | patients with CP and AT | patients with CP and T2D | ||
n | 1529 | 2394 | 136 | 258 | |
Age, years | 18–70 | 18–64 | 40–79 | 26–62 | |
Sex, % | male | 20–80 | 19–73 | 5–80 | 19–51 |
female | 20–80 | 27–81 | 20–95 | 49–81 | |
Smokers, % | 0–50 | 17–21 | 24–56 | 17–52 | |
Depth of periodontal pockets, mm | < 3 | ≥ 5 | ≥ 5 | ≥ 5 | |
Bleeding index, % | < 10 | ≥ 30 | ≥ 30 | ≥ 30 | |
Glycosylated hemoglobin level, % | < 6% | < 6 | < 7% | > 6,5 | |
The peculiarity of the comparative data contained in all 6 sources on the association of CP and AT is the study of not only the subgingival microbiome, but also the taxonomic composition of bacteria found in AT plaques of the vessel walls surgically removed in the process of providing both emergency and routine care to patients.
Thus, not only by metagenomic methods but also by PCR, it was found that first-order periodontal pathogenic bacteria belonging to the P. gingivalis, A. actinomycetemcomitans, and T. forsythia species were detected in more than 60% of samples obtained after endarterioectomy in Spanish patients [43]. However, PCR data of blood clots from Japanese patients with acute myocardial infarction showed that among periodontal pathogens, A. actinomycetemcomitans was the most common organism, detected in approximately 20% of samples [44].
Furthermore, AT plaques contained Acinetobacter (39%), Chryseobacterium (9%), Rhizobium (5%) and Staphylococcus (4%). Comparison of this microbiome with the subgingival microbiota showed that 22 bacterial genera were common to the two different loci, with Acinetobacter appearing to be the most prevalent [40].
In another study, bacterial DNA was detected in 12 endarteriectomy specimens (34.3%). Twenty-three bacterial species/phylotypes were identified. Proteobacteria and Firmicutes accounted for 78.3% and 21.7% of the identified taxa, respectively. However, 15 (60.9%) taxa belonged to bacteria not cultured on nutrient media. The periodontal pathogen A. actinomycetemcomitans was detected in 7 (20%) samples, followed by species from the Pseudomonas genus. Thus, the results suggest a role for the oral microbiota, particularly A. actinomycetemcomitans, in the development of inflammation during atherogenesis [45].
It is important to note that in no case the presence of representatives of F. alocis species in AT plaques was indicated; therefore, these data on the microbiome of AT plaques were not included in the ongoing meta-analysis.
Comparative analysis of the taxonomic composition of the subgingival microbiome in the study groups
First of all, the ratio of the following bacteria belonging to different types was determined for comparative assessment of general trends of changes in the microbiome in groups of people with healthy periodontium, CP without systemic pathology, CP in association with AT or with T2D: Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, Fusobacteria, Synergistes, Spirochaetes.
Comparative statistical analysis of the results was carried out by one-factor analysis of variance (One-way ANOVA) and was aimed at investigating the significance of the difference between the mean values taking into account the scatter (variance) of the data for each indicator. The Fisher's criterion (F) was used as a quantitative expression of such variance, and the probability (p) of its statistical significance was indicated.
As follows from the presented data, statistically significant differences in the percentage of individual types of bacteria in the microbiome, except for the taxonomic group TM7, were not found (Table 4) due to the high heterogeneity of the data. This conclusion also applies to the Firmicutes type, to which the main object of our attention belongs — the F. alocis species.
Table 4. Representation of bacteria of each type in the subgingival microbiome by study group
Bacteria type | Bacterial representation in the microbiome, % (median (minimum; maximum)) | One-way ANOVA | ||||
healthy patients | patients with CP | patients with CP and AT | patients with CP and T2D | F | p | |
Firmicutes | 29,0 (0,6; 43,3) | 30,2 (1,1; 41,0) | 45,6 (38,3; 52,8) | 31,4 (24,5; 68,8) | 2,162 | 0,126 |
Proteobacteria | 30,2 (15,4; 57,9) | 27,9 (15,5; 29,2) | 18,6 (15,1; 22,2) | 17,2 (6,5; 26,6) | 0,958 | 0,438 |
Bacteroides | 10,4 (0; 31,0) | 11,7 (0,1; 35,0) | 17,5 (5,7; 11,0) | 18,8 (11,8; 28,5) | 0,964 | 0,426 |
Actinobacteria | 15,6 (3,6; 16,2) | 7,3 (5,7; 11,0) | 2,7 (1,5; 3,9) | 15,4 (7,2; 16,9) | 1,211 | 0,340 |
Fusobacteria | 12,6 (2,0; 15,6) | 16,4 (8,0; 18,7) | 7,4 (5,1; 9,7) | 9,8 (3,1; 17,0) | 0,936 | 0,448 |
TM7 | 1,3 (0; 1,8) | 4,2 (2,5; 6,5) | – | 1 (0; 2,1) | 7,879 | 0,013 |
Spirochaetes | 0,9 (0; 2,7) | 2,3 (0,1; 20,0) | 6,1 (5,3; 7,0) | 0,4 (0; 1,1) | 0,799 | 0,522 |
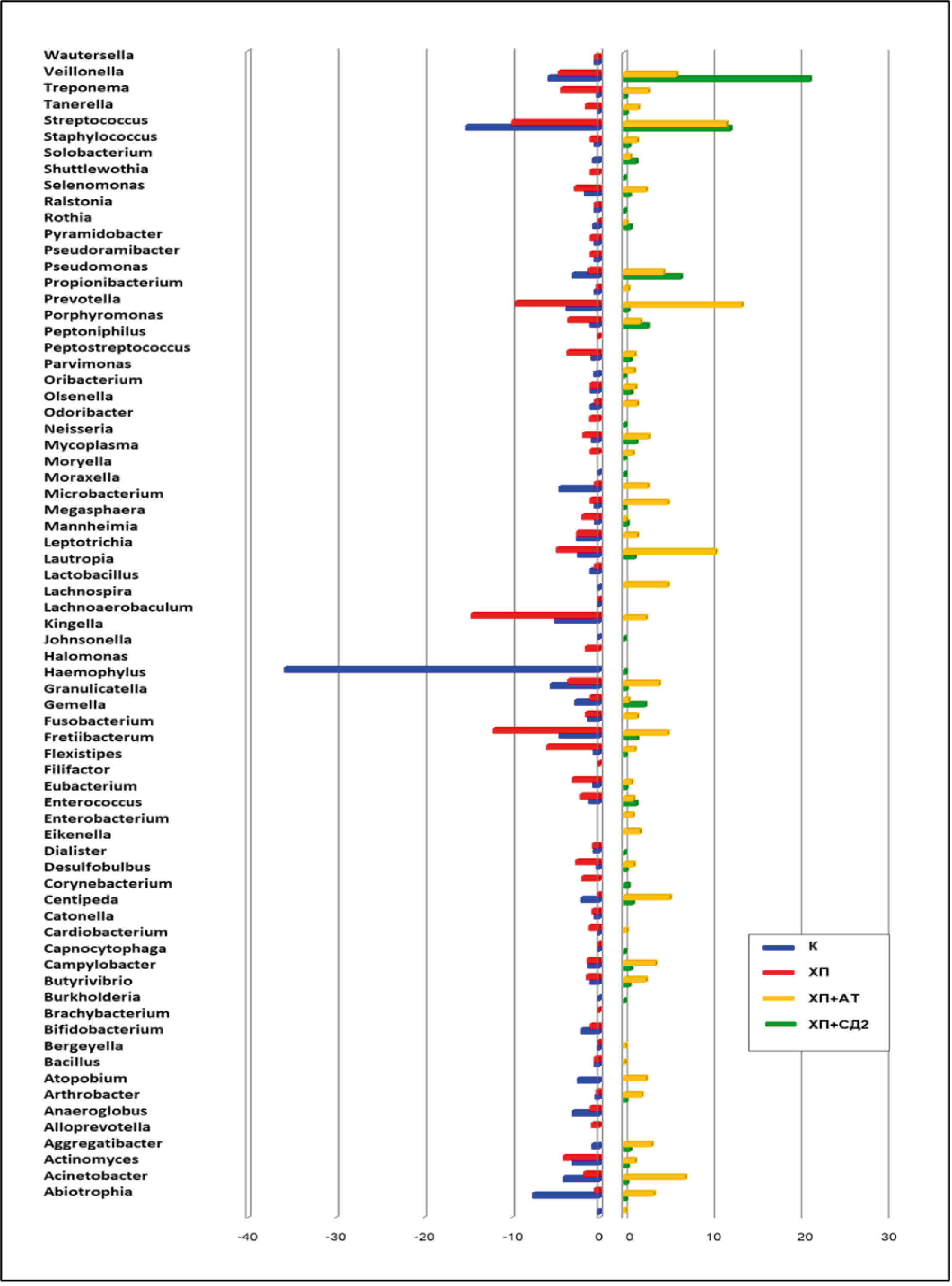
The results obtained in a comparative aspect concerned the percentage of individual bacteria of different genera in different study groups (Fig. 1).
Fig. 1. Representation of the most frequent bacterial genera in the subgingival microbiome by study group.
Analysis of the obtained data shows that in all samples of the subgingival microbiome containing representatives of the Filifactor genus and included in this study, this genus was accompanied by another 70 genera of bacteria. In those cases, where the literature source had data on the species composition of the microbiome, the Filifactor genus included only one species – F. alocis, and the content of Filifactor genus bacteria was different in different study groups. Thus, in the group of healthy people the share of these bacteria (in %) among representatives of other species with the designation of median (minimum; maximum) was only 0.66 (0.1; 5.8); in the group of CP without systemic pathology — 3 (0.1; 13.0); in the association of CP + AT the index was also low — 0.12 (0.05; 0.27); and in the association of CP + T2D — significantly higher: 1.6 (1.0; 2.3).
Thus, Filifactor genus bacteria showed a relatively low content in the composition of the subgingival microbiome in healthy subjects and in the association of CP + AT. In the association of CP + T2D, the median proportion of these bacteria in the periodontal microbiome was 2.4 times higher than in healthy subjects, and in patients with CP without systemic pathology — 4.5 times higher. Nevertheless, the scatter of data for each sample did not allow to establish statistically significant differences when comparing by One-way ANOVA: in the groups "healthy individuals - CP", F = 1.910 at p = 0.192; in the groups "healthy individuals — CP + AT", F = 0.427 at p = 0.527; in the groups "healthy individuals — CP + T2D", F = 0.002 at p = 0.963.
Representatives of the following genera predominated in the subgingival microbiome of healthy people: Haemophilus, Streptococcus, Abiotrophia, Granulicatella, Moraxella, Veillonella; in patients with CP without systemic pathology — Fusobacterium, Fretibacterium, Lachnoaerobaculum, Prevotella, Streptococcus; in CP + AT — Streptococcus, Veillonella, Propionibacterium; in CP + T2D — Prevotella, Streptococcus, Leptotrix, Veillonella, Actinomyces.
All these data served as a basis for correlation analysis, which allowed us to determine possible relationships of bacteria of the genus Filifactor with other bacteria of the same localization.
Meta-analysis of correlations of bacteria of the genus Filifactor with other bacterial genera in the subgingival microbiome in the study groups
Correlation analysis of the data in the form of pairwise correlations was performed by determining the Spearman correlation coefficient (r) and its statistical significance (p) between bacteria of the genus Filifactor (species F. alocis) and other bacteria in the subgingival microbiome separately for each study group.
The content of bacteria of the genus Filifactor, represented by only one species of F. alocis, in the composition of the gingival sulcus microbiome of people with healthy periodontium was relatively small and was correlated at a statistically significant level with bacteria of 8 genera (Fig. 2, a). The correlations were positive with bacteria of the genera Actinomyces, Brachybacterium, and Mogibacterium, which, like Filifactor, are Gram-positive microorganisms, while the correlations were negative with the other 5 genera (Acinetobacter, Alloprevotella, Campylobacter, Misrobacterium, and Moraxella).
Fig. 2. Correlations of F. alocis with other bacteria of the subgingival microbiome in healthy subjects (a), in CP (b), CP + AT (c), CP + T2D (d). DOI: https://doi.org/10.36233/0372-9311-500-2
The diameter of the circle denoting the genus of the bacterium roughly reflects the quantitative representation of this genus of bacteria among the other detected genera. Solid arrows indicate positive correlation, dashed arrows indicate negative correlation. The presented figures show only statistically significant correlations at p < 0.05.
The content of bacteria of Filifactor genus in complication of CP with such systemic disease as AT was minimal (Fig. 2, c). Nevertheless, the abundance of correlation pairs, the growth of the spectrum of microorganisms involved in them with a clear predominance of positive correlations draws attention. Such bacterial genera as Aggregatibacter, Atopobium, Cardiobacterium, Fretibacterium, Haemophylus, Halomonas, Kingella, Megasphera, Peptostreptococcus, Streptococcus, Treponema, Veillonella appear in the composition of correlation pairs, among which only 3 genera (Atopobium, Peptostreptococcus, Streptococcus) belong to Gram-positive microorganisms.
Fig. 2, d shows the range of correlations of F. alocis, whose abundance was about 10 times higher than in concomitant AT, with the bacterial genera included in the subgingival microbiome in CP associated with T2D. In the correlation analysis in this aspect, the number of pairwise correlations fell compared to the other pathological conditions analyzed to 11, with approximately the same ratio of positive to negative correlations. Among positive correlations we can note the participation of Gram-negative bacteria of the genera Aggregatibacter, Alloprevotella, Dialister, and among Gram-positive bacteria — Atopobium, Eubacterium, Lachnoanaerobaculum.
Discussion
Evaluating the results of correlation analysis, several features should be emphasized. In all groups, the content of F. alocis bacteria was different – from minimal in the association of CP and AT to maximum in CP without systemic effects. In the composition of correlation pairs there was not a single genus of bacteria that occurred in all 4 groups. The diagrams presented in our work indicate the existence of numerous correlations between F. alocis and other components of the microbiome, which, in combination with the data on the presence of its exotoxin, as well as A. actinomycetemcomitans [12], allows us to raise the question of its classification as a periodontal pathogenic species of the first order.
One of the foreign meta-analyses conducted within the framework of the international Human Microbiome Project showed that supra- and sub-gingival dental deposits (biofilm) are represented by 14 taxa of microorganisms, which are the most numerous both in terms of frequency of detection and quantitative expression in the composition of gingival biofilm: Actinomyces, Aggregatibacter, Capnocytophaga, Corynebacterium, Haemophilus, Fusobacterium, Neisseria, Prevotella, Porphyromonas, Rothia, Selenomonas, Streptococcus, Veillonella, Wolinella [6].
Based on the purpose of the meta-analysis presented in our article, special attention should be paid to those bacteria that appeared in the correlation pairs: either only in CP without systemic effects, only in the association of CP with both AT and T2D, only in AT or only in T2D. The species affiliation of the bacteria associated correlatively with F. alocis could not be determined because only a few of the selected literature sources contained this information, which was insufficient to identify statistically significant correlations. Only the first category of correlations (CP without systemic effects) with Filifactor genus bacteria included representatives of 8 genera: Desulfobulbus, Gemella, Granulicatella, Lachnospira, Mycoplasma, Odoribacter, Oribacterium, Pseudoramibacter. Among the genera mentioned, there were no bacterial genera containing generally recognized periodontal pathogens. The second category associated with the manifestation of systemic effects of periodontal diseases included only 3 genera: Aggregatibacter, Atopobium, Megasphera. Representatives of the Aggregatibacter genus, which was included in the composition of statistically significant correlation pairs, were registered in 58% of samples, and the other 2 genera — in 42%. In those sources that contained information on the species composition of bacteria in the subgingival microbiome, bacteria of the genus Aggregatibacter included only one species — A. actinomycetemcommitans, which is classified as periodontal pathogen of the first order.
Only at AT correlation significant genera were registered: Cardiobacterium, Fretibacterium, Haemophylus, Halomonas, Kingella, Leptotrichia, Peptostreptococcus, Streptococcus, Treponema, Veillonella. Only at T2D, 4 genera were noted in the composition of correlative pairs: Dialister, Lachnoanaerobaculum, Rothia, Sphingomonas. In terms of species composition, where indicated, affiliation with periodontal pathogens was not noted in any case.
Conclusion
- A comparative meta-analysis of the subgingival microbiome under normal circumstances, in regular CP and CP associated with comorbid pathology, covering 1529 healthy and 2394 patients with CP was carried out, which allowed to obtain fundamentally new data confirming the differences of comparison groups in the composition of the microbiome. According to taxonomic characterization, representatives of the following genera prevailed in the composition of the subgingival microbiome of healthy people: Haemophilus, Streptococcus, Abiotrophia, Granulicatella, Moraxella, Veillonella; in patients with CP without systemic pathology — Fusobacterium, Fretibacterium, Lachnoaerobaculum, Prevotella, Streptococcus; in CP in association with AT — Streptococcus, Veillonella, Propionibacterium; in CP in association with T2D — Prevotella, Streptococcus, Leptotriсhia, Veillonella, Actinomyces.
- As a result of the meta-analysis of the data on the genus of bacteria that are correlated with Filifactor genus bacteria, which in those cases when the sources contained information on the species composition of bacteria, belonged to the F. alocis genus, it was found that the development of CP, conditionally not associated with systemic pathology, did not allow to determine significant correlations of these bacteria with other periodontal pathogens. At the same time, the place of F. alocis as a periodontal pathogenic species (probably of the first order) was clarified, since it is practically not found in the normal microbiota, indicating its relatively high virulence.
- In groups of patients in whom CP was accompanied by the presence of systemic pathology — AT or T2D, in addition to associates-commensals, the presence of statistically significant correlations with the genus of Aggregatibacter bacteria containing periodontal pathogenic species of the first order — A. actinomycetemcomitans was confirmed. Representatives of the Porphyromonas (P. gingivalis) and Tannerella (T. forsythia) genera seem to be of independent importance as periodontal pathogenic species of the first order, due to their high virulence properties, regardless of symbionts from other taxa of the subgingival microbiome.
1 National Center for Biotechnology Information. Taxonomy Browser. URL: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi
About the authors
Tatyana V. Tsareva
Russian University of Medicine
Email: nikola777@rambler.ru
ORCID iD: 0000-0001-9571-0520
Cand. Sci. (Med.), Associate Professor, Department of microbiology, virology, immunology
Russian Federation, MoscowIrina P. Balmasova
Russian University of Medicine
Email: nikola777@rambler.ru
ORCID iD: 0000-0001-8194-2419
D. Sci. (Med.), Professor, Head, Laboratory of pathogenesis and methods of treatment of infectious diseases, Scientific Research Medical Dental Institute
Russian Federation, MoscowVictor N. Tsarev
Russian University of Medicine
Author for correspondence.
Email: nikola777@rambler.ru
ORCID iD: 0000-0002-3311-0367
D. Sci. (Med.), Professor, Head, Department of microbiology, virology, immunology
Russian Federation, MoscowReferences
- Кузьмина Э.М., Янушевич О.О., Кузьмина И.Н. Стоматологическая заболеваемость населения России. Эпидемиологическое стоматологическое обследование. М.;2019. Kuz'mina E.M., Yanushevich O.O., Kuz'mina I.N. Dental Morbidity of the Russian Population. Epidemiological Dental Examination. Moscow;2019.
- Abusleme L., Dupuy A.K., Dutzan N., et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7(5): 1016–25. DOI: https://doi.org/10.1038/ismej.2012.174
- Kassebaum N.J., Bernabé E., Dahiya M., et al. Global burden of severe periodontitis in 1990–2010: a systematic review and meta-regression. J. Dent. Res. 2014;93(11):1045–53. doi: https://doi.org/10.1177/0022034514552491
- Könönen E., Gursoy M., Gursoy U.K. Periodontitis: a multifaceted disease of tooth-supporting tissues. J. Clin. Med. 2019;8(8):1135. DOI: https://doi.org/10.3390/jcm8081135
- Tonetti M.S., Jepsen S., Jin L., Otomo-Corgel J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: a call for global action. J. Clin. Periodontol. 2017;44(5):456–62. doi: https://doi.org/10.1111/jcpe.12732
- Балмасова И.П., Царев В.Н., Янушевич О.О. и др. Микроэкология пародонта. Взаимосвязь локальных и системных эффектов. М.;2021. Balmasova I.P., Tsarev V.N., Yanushevich O.O., et al. Microecology of Periodontal Disease. The Relationship of Local and Systemic Effects. Moscow;2021. EDN: https://elibrary.ru/myzmbu
- Bui F.Q., Almeida-da-Silva C.L.C., Huynh B., et al. Association between periodontal pathogens and systemic disease. J. Biomed. Sci. 2019;42(1):27–35. doi: https://doi.org/10.1016/j.bj.2018.12.001
- Царев В.Н., Николаева Е.Н., Ипполитов Е.В. Пародонтопатогенные бактерии – основной фактор возникновения и развития пародонтита. Журнал микробиологии, эпидемиологии и иммунобиологии. 2017;(5):101–12. Tsarev V.N., Nikolaeva E.N., Ippolitov E.V. Periodontophatogenic bacteria of the main factors of emergence and development of periodontitis. Journal of Microbiology, Epidemiology and Immunobiology. 2017;(5):101–12. doi: https://doi.org/10.36233/0372-9311-2017-5-101-112 EDN: https://elibrary.ru/ctbcar
- Nikolaeva E.N., Tsarev V.N., Tsareva T.V., et al. Interrelation of cardiovascular diseases with anaerobic bacteria of subgingival biofilm. Contemp. Clin. Dent. 2019;10(4):637–42. doi: https://doi.org/10.4103/ccd.ccd_84_19
- Socransky S.S., Haffajee A.D., Cugini M.A., et al. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998;25(2):134–44. doi: https://doi.org/10.1111/j.1600-051x.1998.tb02419.x
- Ahmed H.J., Svensson J.A., Cope L.D., et al. Prevalence of cdtABC genes encoding cytolethaldistanding toxin among Haemophilus ducreyi and Actinobacillus actinomycetemcomitans strains. J. Med. Microbiol. 2001;50(10):860–4. doi: https://doi.org/10.1099/0022-1317-50-10-860
- Ozuna H., Snider I., Belibasakis G.N., et al. Aggregatibacter actinomycetemcomitans and Filifactor alocis: Two exotoxin-producing oral pathogens. Front. Oral Health. 2022;3:981343. DOI: https://doi.org/10.3389/froh.2022.981343
- Ушаков Р.В., Царев В.Н. Антимикробная терапия в стоматологии: принципы и алгоритмы. М.;2019. Ushakov R.V., Tsarev V.N. Antimicrobial Therapy in Dentistry: Principles and Algorithms. Moscow;2019.
- Rafiei M., Kiani F., Sayehmiri K., et al. Prevalence of anaerobic bacteria (P. gingivalis) as major microbial agent in the incidence periodontal diseases by meta-analysis. J. Dent. (Shiraz). 2018;19(3):232–42.
- Hiranmayi K.V., Sirisha K., Ramoji Rao M.V., Sudhakar P. Novel pathogens in periodontal microbiology. J. Pharm. Bioallied Sci. 2017;9(3):155–63. doi: https://doi.org/10.4103/jpbs.jpbs_288_16
- Moffatt C.E., Whitmore S.E., Griffen A.L., et al. Filifactor alocis interactions with gingival epithelial cells. J. Mol. Oral Microbiol. 2011;26(6):365–73. doi: https://doi.org/10.1111/j.2041-1014.2011.00624.x
- Wang Q., Wright C.J., Dingming H., et al. Oral community interactions of Filifactor alocis in vitro. PLoS One. 2013;8(10):e76271. DOI: https://doi.org/10.1371/journal.pone.0076271
- Aja Е., Mishra А., Dou Н., Fletcher H.M. Role of the Filifactor alocis hypothetical protein FA519 in oxidative stress resistance. Microbiol. Spectr. 2021;9(3):e0121221. doi: https://doi.org/10.1128/spectrum.01212-21
- Царева Т.В., Янушевич О.О., Царев В.Н., Балмасова И.П. Бактерии рода Filifactor у больных пародонтитом и сахарным диабетом по данным метагеномного анализа микробиома пародонта. Журнал микробиологии, эпидемиологии и иммунобиологии. 2023;100(6):485–94. Tsareva T.V., Yanushevich O.O., Tsarev V.N., Balmasova I.P. Bacteria of genus Filifactor in patients with periodontitis and type 2 diabetes in accordance with metagenomic analysis of the periodontal microbiome. Journal of Microbiology, Epidemiology and Immunobiology. 2023;100(6):485–94. doi: https://doi.org/10.36233/0372-9311-428 EDN: https://elibrary.ru/yqoxxk
- Янушевич О.О., Царёв В.Н., Балмасова И.П. и др. Первый опыт применения отечественного диагностического набора генетических праймеров для выявления нового пародонтопатогена Filifactor alocis и его ассоциации с Porphyromonas gingivalis. Клиническая лабораторная диагностика. 2022;67(12):744–8. Yanushevich O.O., Tsarev V.N., Balmasova I.P., et al. The first experience of using a domestic diagnostic set of genetic primers to identify a new periodontopathogen Filifactor alocis and its association with Porphyromonas gingivalis. Clinical Laboratory Diagnostics. 2022;67(12):744–8. DOI: https://doi.org/10.51620/0869-2084-2022-67-12-744-748 EDN: https://elibrary.ru/djlzfu
- Янушевич О.О., Царев В.Н., Николаева Е.Н. и др. Первый отечественный опыт выявления ассоциации анаэробных бактерий Filifactor alocis и Porphyromonas gingivalis молекулярно-биологическими методами при заболеваниях пародонта и коморбидной патологии (сравнительное исследование). Вестник Российской академии медицинских наук. 2022;77(6):437–46. Yanushevich O.O., Tsarev V.N., Nikolaeva E.N., et al. The first domestic experience of detecting the association of anaerobic bacteria Filifactor alocis and Porphyromonas gingivalis by molecular biological methods in periodontal diseases and comorbid pathology (comparative research). Annals of the Russian Academy of Medical Sciences. 2022;77(6):437–46. DOI: https://doi.org/10.15690/vramn2262 EDN: https://elibrary.ru/pzajok
- Kumar P.S., Griffen A.L., Moeschberger M.L., et al. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 2005;43(8):3944–55. doi: https://doi.org/10.1128/jcm.43.8.3944-3955.2005
- Griffen A.L., Beall C.J., Campbell J.H., et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6(6):1176–85. DOI: https://doi.org/10.1038/ismej.2011.191
- Liu B., Faller L.L., Klitgord N., et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7(6):e37919. doi: https://doi.org/10.1371/journal.pone.0037919
- Costalonga M., Herzberg M.C. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 2014;162(2 Pt. A):22–38. doi: https://doi.org/10.1016/j.imlet.2014.08.017
- Filho W.S.S., Casarin R.C.V., Junior E.L.N., et al. Microbial diversity similarities in periodontal pockets and atheromatous plaques of cardiovascular disease patients. PLoS One. 2014;9(10):e109761. doi: https://doi.org/10.1371/journal.pone.0109761
- Lourenço T.G.B., Heller D., da Silva-Boghossian C.M., et al. Microbial signature profiles of periodontally healthy and diseased patients. J. Clin. Periodontol. 2014;41(11):1027–36. doi: https://doi.org/10.1111/jcpe.12302
- Pérez-Chaparro P.J., Gonçalves C, Figueiredo L.C., et al. Newly identified pathogens associated with periodontitis. J. Dent. Res. 2014;93(9):846–58. doi: https://doi.org/10.1177/0022034514542468
- Park O.J., Yi H., Jeon J.H., et al. Pyrosequencing analysis of subgingival microbiota in distinct periodontal conditions. J. Dent. Res. 2015;94(7):921–7. doi: https://doi.org/10.1177/0022034515583531
- Ziganshina E.E., Sharifullina D.M., Lozhkin A.P., et al. Bacterial communities associated with atherosclerotic plaques from Russian individuals with atherosclerosis. PLoS One. 2016;11(10):e0164836. doi: https://doi.org/10.1371/journal.pone.0164836
- Demmer R.T., Breskin A., Rosenbaum M., et al. The subgingival microbiome, systemic inflammation and insulin resistance: The oral infections, glucose intolerance and insulin resistance study. J. Clin. Periodontol. 2017;44(3):255–65. doi: https://doi.org/10.1111/jcpe.12664
- Ganesan S.M., Joshi V., Fellows M., et al. A tale of two risks: smoking, diabetes and the subgingival microbiome. ISME J. 2017;11(9):2075–89. doi: https://doi.org/10.1038/ismej.2017.73.
- Long J., Cai Q., Steinwandel M., et al. Association of oral microbiome with type 2 diabetes risk. J Periodontal Res. 2017;52(3):636–43. https://doi.org/10.1111/jre.12432
- Graves D.T., Corrêa J.D., Silva T.A. The oral microbiota is modified by systemic diseases. J. Dent. Res. 2019;98(2): 148–56. DOI: https://doi.org/10.1177/0022034518805739
- Rodríguez-Hernández A.P., de Lourdes Márquez-Corona M., Pontigo-Loyola A.P., et al. Subgingival microbiota of Mexicans with type 2 diabetes with different periodontal and metabolic conditions. Int. J. Environ. Res. Public Health. 2019;16(17):3184. doi: https://doi.org/10.3390/ijerph16173184
- Saeb A.T.M., Al-Rubeaana K.A., Aldosaryb K., et al. Relative reduction of biological and phylogenetic diversity of the oral microbiota of diabetes and pre-diabetes patients. Microb. Pathog. 2019;128:215–29. doi: https://doi.org/10.1016/j.micpath.2019.01.009
- Matsha T.E., Prince Y., Davids S., et al. Oral microbiome signatures in diabetes mellitus and periodontal disease. J. Dent. Res. 2020;99(6):658–65. doi: https://doi.org/10.1177/0022034520913818
- Bouzid F., Gtif I., Alfadhli S., et al. A potential oral microbiome signature associated with coronary artery disease in Tunisia. Biosci. Rep. 2022;42(7):BSR20220583. doi: https://doi.org/10.1042/BSR20220583
- Curia M.C., Pignatelli P., D’Antonio D.L., et al. Oral Porphyromonas gingivalis and Fusobacterium nucleatum abundance in subjects in primary and secondary cardiovascular prevention, with or without heterozygous familial hypercholesterolemia. Biomedicines. 2022;10(9):2144. doi: https://doi.org/10.3390/biomedicines10092144
- Rao A., Lokesh J., D'Souza C., et al. Metagenomic analysis to uncover the subgingival and atherosclerotic plaque microbiota in patients with coronary artery disease. Indian J. Microbiol. 2023;63(3):281–90. doi: https://doi.org/10.1007/s12088-023-01082-9
- Kato-Kogoe N., Sakaguchi S., Kamiya K., et al. Characterization of salivary microbiota in patients with atherosclerotic cardiovascular disease: а case-control study. J. Atheroscler. Thromb. 2022;29(3):403–21. doi: https://doi.org/10.5551/jat.60608
- Solbiati J., Frias-Lopez J. Metatranscriptome of the oral microbiome in health and disease. J. Dent. Res. 2018;97(5):492–500. DOI: https://doi.org/10.1177/0022034518761644
- Figuero E., Sánchez-Beltrán M., Cuesta-Frechoso S., et al. Detection of periodontal bacteria in atheromatous plaques by nested polymerase chain reaction. J. Periodontol. 2011;82(10): 1469–77. DOI: https://doi.org/10.1902/jop.2011.100719
- Ohki T., Itabashi Y., Kohno T., et al. Detection of periodontal bacteria in thrombi of patients with acute myocardial infarction by polymerase chain reaction. Am. Heart J. 2012;163(2):164–7. DOI: https://doi.org/10.1016/j.ahj.2011.10.012
- Calandrini C.A., Ribeiro A.C., Gonnelli A.C., et al. Microbial composition of atherosclerotic plaques. Oral Dis. 2014; 20(3):128–34. DOI: https://doi.org/10.1111/odi.12205
Supplementary files