Genotypic portrait of SARS-CoV-2 in Primorsky Krai during the COVID-19 pandemic
- Authors: Popova A.Y.1, Shchelkanov M.Y.2,3, Krylova N.V.2,3, Belik A.A.2, Semeikina L.M.2,4, Zaporozhets T.S.2, Smolenskiy V.Y.1, Persianova E.V.2, Prosyannikova M.N.4, Belov Y.A.2,3, Iunikhina O.V.2,3, Pott A.B.2, Khomichuk T.F.4, Simakova A.I.5, Abramova S.A.2, Romanova O.B.4, Detkovskaya T.N.6, Kryzhanovskiy S.P.7, Besednova N.N.2,7
-
Affiliations:
- Federal Service for the Oversight of Consumer Protection and Welfare
- G.Р. Somov Institute of Epidemiology and Microbiology
- Far Eastern Federal University
- Center for Hygiene and Epidemiology in Primorsky Krai
- Pacific State Medical University
- Office of the Federal Service for the Oversight of Consumer Protection and Welfare for the Primorsky Krai
- Medical Association of the Far Eastern Branch of the Russian Academy of Sciences
- Issue: Vol 101, No 1 (2024)
- Pages: 19-35
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18530
- DOI: https://doi.org/10.36233/0372-9311-497
- EDN: https://elibrary.ru/pujffa
- ID: 18530
Cite item
Abstract
Introduction. The COVID-19 pandemic, etiologically related to SARS-CoV-2, was the longest-lasting pandemic for an acute respiratory disease and had a significant impact on demography, economics and politics globally. Experiences with this pandemic are significant for the sustainable development of human society. A detailed analysis of these experiences in epidemic control should include details of the pathogen evolution down to the regional level.
The aim of the study was to establish the regularities of the COVID-19 epidemic process in connection with the change of the pathogen genetic variants on the territory of Primorsky Krai.
Materials and methods. A retrospective epidemiological analysis of COVID-19 incidence and dynamics of circulation of SARS-CoV-2 genetic variants during the pandemic of this disease (11.03.2020–05.05.2023) in Primorsky Krai was carried out. Data was gathered from the Department of Rospotrebnadzor in Primorsky Krai, the Centre of Hygiene and Epidemiology in Primorsky Krai, as well as the G.P. Somov Research Institute of Epidemiology and Microbiology of the Russian Federation and Russian Platform for Aggregation of Information on Virus Genomes (VGARus). In particular, 1055 nucleotide sequences of full-length SARS-CoV-2 genomes from Primorsky Krai, among which 553 were sequenced at the G.P. Somov Research Institute of Epidemiology and Microbiology were included in analysis.
Results. When analyzing the epidemic dynamics of COVID-19 in Primorsky Krai (2020–2023), 7 rises in incidence with different clinical and epidemiological symptoms depending on the genetic variants of the pathogen were identified. At the beginning of the pandemic in Primorsky Krai, as well as throughout Russia, Wuhan-like variants of SARS-CoV-2 were predominant, though later, Delta and Omicron genetic variants were in the majority. By the end of April — beginning of May 2023, the proportion of Omicron sub-variants (XBB.1.9.2 and XBB.1.16) in Primorsky Krai was higher than the Russian average and comparable to that in neighboring countries (Republic of Korea and Japan).
Conclusion. Due to the ongoing evolution of SARS-CoV-2, the possibility of the emergence of new pathogens, the peculiarities of the geographical location as well as political and economic importance of Primorsky Krai, it is necessary to consistently improve regional capabilities for operational molecular virological monitoring.
Full Text
Introduction
Coronavirus disease 2019 (COVID-19) is etiologically related to Severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2; Nidovirales: Coronaviridae, Betacoronavirus, subgenus Sarbecovirus) [1, 2] and resulted from the pathogen crossing the interspecies barrier and entering the human population from bat populations (Chiroptera, Microchiroptera) [3, 4]. COVID-19 caused the longest (11.03.2020–05.05.2023, or 1150 days), largest (over 750 million confirmed cases) and one of the deadliest (about 6 million deaths) acute respiratory disease pandemic in human history [3, 5]. The COVID-19 pandemic was the first to be associated with a coronavirus of bats (Chiroptera) [1, 6], the second largest mammalian group after rodents, with a number of unique evolutionary features [7–9], the epidemiological significance of which became clear only in the early 21st century after the epidemics caused by SARS coronaviruses (Nidovirales: Coronaviridae, Betacoronavirus, subgenus Sarbecovirus) [1, 10] and Middle East respiratory syndrome (Nidovirales: Coronaviridae, Betacoronavirus, subgenus Merbecovirus) [11, 12].
The COVID-19 pandemic stimulated the widespread introduction of molecular genetic [13, 14] and information [15, 16] technologies into the practice of anti-epidemic measures. With the degradation of broad international relations in the early 2020s, exacerbated by the pandemic, and the declining authority of international organizations, including the World Health Organization (WHO), the Russian Federation was forced to deploy its own national molecular genetic data platform, the Virus genome aggregator of Russia (VGARus), developed by the Federal Service for Supervision of Consumer Rights Protection and Human Welfare (Rospotrebnadzor). VGARus was put into operation at the Central Research Institute of Epidemiology of Rospotrebnadzor by the Order No. 448 of the Russian Government from 23.03.2021. By the end of 2023, more than 150 organizations were registered in the system, carrying out its replenishment; the database contains more than 300 thousand nucleotide sequences of SARS-CoV-2, more than half of which are full-length viral genomes [14, 17]. Thus, unlike even the last influenza A (H1N1 pdm09) (Articulavirales: Orthomyxoviridae, Alphainfluenzavirus) pandemic (2009–2010) [18], when molecular genetic methods were actively used to monitor the infectious process, the COVID-19 pandemic was the first global epidemic process that was characterized in detail using real-time whole-genome sequencing data (at least in developed countries) [14, 17, 19].
Russia, being the largest territory in the world, has an enormous regional diversity of natural, climatic and socio-demographic conditions, which should be taken into account in the process of epidemiological analysis of infectious disease dynamics. For example, Primorsky Krai (PK), located in the south of the Russian Far East (practically at the latitude of the Crimean Peninsula), is characterized by a unique monsoon temperate climate formed by a latitudinal climatic anomaly due to the cold Primorsky Current flowing through from the Tatar Strait. The PK provides convenient access via non-freezing ports (Vladivostok, Vostochny, Zarubino, Nakhodka, Posyet) to the dynamically developing Asia-Pacific region and is traditionally closely connected with the north-eastern provinces of the People's Republic of China by all means of transport. The administrative center of the PK, Vladivostok, being the capital of the Far Eastern Federal District, is well recognized both in Russia and abroad due to its free port status and the presence of the Far Eastern Federal University, one of the largest educational facilities in the Asian part of Russia.
The aim of this study is to analyze the regularities of the COVID-19 epidemic process development in the PK territory during the pandemic period of this disease with the help of molecular genetics (2020–2023).
Materials and methods
Nasopharyngeal swabs for PCR testing and obtaining viral strains were collected from people living in the PK territory. Biological material was collected by qualified personnel who had undergone preliminary training and regular skills testing on rhinolaryngological simulators [20, 21]. The study was conducted with the voluntary informed consent of the patients. The study protocol was approved by the Ethical Committee of the G.Р. Somov Institute of Epidemiology and Microbiology (protocol No. 2 of November 11, 2021).
SARS-CoV-2 RNA was detected by reverse transcription followed by quantitative polymerase chain reaction (RT-qPCR): RNA isolation was performed manually using the M-Sorb-NK reagent complex (Syntol) or (in case of large amounts of samples) at the AutoPure 96 automatic station (Hangzhou Allsheng Instruments Co.), RT-PCR-SARS-CoV-2 reagent kit (Syntol). Positive samples intended for subsequent use for research purposes were stored at –20ºC while maintaining the cold chain.
Sequencing of full-length SARS-CoV-2 genome was performed for positive samples with threshold cycle in primary PCR testing Ct ≤ 25 on the Nanopore technology platform [22, 23] according to ARTIC SARS-CoV-2 v3 protocol: reverse transcription was performed using the Midnight RT-PCR Expansion kit; amplicons were obtained and coded using 29 pairs of overlapping primers and the Rapid Barcoding Kit 96 (SQK-RBK110. 96); cDNA purification was performed on AMPure XP beads magnetic particles; the resulting genomic libraries were sequenced on a MinION instrument using FLO-MIN106 R9.4.1 cells (Oxford Nanopore Technologies). The obtained data in FAST5 format (the MinKNOW software package was converted to FASTQc format using the Guppy v. 6.3.8 program). SARS-CoV-2 genomes were assembled in FASTA format by alignment with reference sequences from VGARus using the Epi2me v. 22 software package and the ARTIC v. 1 module. To assess the quality of the assembled sequences and genome distribution among lineages, the Nextclade and Pangolin COVID-19 Lineage Assigner v. 4.3 services were used. Sequences in which the number of unrecognized or ambiguous nucleotides was more than 10% of the full genome sequence of SARS-CoV-2 were excluded from the sample for analysis.
Phylogenetic analysis of nucleotide sequences was performed after multiple alignment using MAFFT v. 7.475 by the "neighbor joining" method [24] using the MEGA v. 11.0.13 software package at the bootstrap support level of 1000 repetitions. Visualization of the obtained data was performed using the iTOL v. 6 service.
Isolation of SARS-CoV-2 strains was performed from qPCR-positive samples of nasopharyngeal washings, clarified by low-speed centrifugation and filtered through Millex (Merck) nozzles with a pore diameter of 0.22 μm, on the Vero E6 cell line model (kidneys of the African green monkey), which was cultured in plastic tubes (Nunc) with a slanted bottom of 5.5 cm2 in DMEM medium (BioloT) supplemented with 1% fetal calf serum (PanEco), 100 U/ml gentamicin at 37ºC in an atmosphere of 5% CO2 [25, 26]. The efficiency of virus isolation was evaluated by decreasing the threshold cycle of PCR in 3 consecutive passages. On the 5th day after infection, the culture fluid was taken for RT-qPCR (after the 3rd passage, we additionally determined lg of the 50% tissue culture infectious dose (lg TCID50) using the MTT test).
The MTT assay was used to assess the survival of Vero E6 cells under the influence of different dilutions of the virus when evaluating the infectious titer of SARS-CoV-2 strains after the 3rd passage. The essence of this method is the ability of viable cells to convert the highly soluble yellow bromide 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) into insoluble intracellular crystals of (E, Z)-5(4,5-dimethylthiazol-2-yl)-1,3-diphenylformazan (MTT-formazan) under the action of intracellular dehydrogenases [27, 28]:
On the 5th day after infection, 20 µl of 5 mg/ml MTT solution (Sigma-Aldrich) was added to the wells with cells in a 96-well plate (Merck); incubated at 37ºC in an atmosphere with 5% CO2 for 2 hours; after removal of culture media, 150 µl of isopropyl alcohol acidified with 0.4 M HCl was added to the wells; the optical density in the well was determined at 540 nm (near the absorption maximum of MTT-formazan) with subtraction of the background value at 620 nm using a tablet reader (Labsystems). The cytopathogenicity level (CPL) of the strain in a particular dilution was calculated according to the following formula:
,
where Dv — optical density of the infected sample; D0 — optical density of uninfected cell culture.
The infectious titre of the strain was defined as the limiting dilution of the initial virus-containing liquid recorded in the experiment, for which CPL > 0 (inequality to zero was assessed by Student's t-test at a significance level of p = 95% and 8-fold repetitions of each dilution of the virus-containing liquid).
Epidemiological analysis of COVID-19 incidence in PK from 20.04.2020 to 30.06.2023 was based on data from the Department of Rospotrebnadzor for Primorsky Krai, the Centre for Hygiene and Epidemiology in Primorsky Krai1, in Russia — on data from the official website of Rospotrebnadzor2 and the Russian information portal Stopcoronavirus3. Information on the distribution of genetic variants of SARS-CoV-2 in Russia was used from the review and analytical work [17, 19] and the Russian database VGARus, in Eastern Asian countries – from the international database GISAID4.
Statistical processing of data was carried out using the following functions from the MS Excel 2010 software package: sample size (n); arithmetic mean (M); standard error of mean (m); median (Me); lower (25%) quartile (LQ); upper (75%) quartile (UQ); interquartile range (LQ-UQ). The normality of distribution of quantitative signs was tested using the Shapiro–Wilk W-test; Student's t-test (at the significance level of the alternative hypothesis p = 0.05); Mann–Whitney U-test [29, 30].
The method of principal components as a way to find a subspace of lower dimensionality, in projection on which the initial data have the smallest spread of quadratic deviations [31, 32], was applied using the software packages "Statistica v. 7.0" ("StatSoft Inc.") and "Mathworks Matlab R2007b" software packages.
Results
Indication of SARS-CoV-2 RNA in nasopharyngeal washings during the COVID 19 pandemic in PK territory was carried out in 24 laboratories (including 5 private laboratories): 3,007,009 RT-qPCR tests were performed, of which 399,821 (13.3%) were positive5. The obtained information was integrated for operational analysis in the epidemiological units of the Centre for Hygiene and Epidemiology in Primorsky Krai and the Primorsky Krai Department of Rospotrebnadzor.
The full-genome nucleotide sequences of SARS-CoV-2 obtained from samples of biological material collected on the territory of the PK during the COVID-19 pandemic are presented in Table 1. In addition to 553 genomes sequenced at the G.P. Somov Research Institute of Epidemiology and Microbiology within the framework of the Federal Project Sanitary Shield of the Country — Safety for Health, another 502 genomes were sequenced in other scientific institutions, to which RT-qPCR-positive samples from the Center of Hygiene and Epidemiology in Primorsky Krai were sent in accordance with the Order of Rospotrebnadzor6.
Table 1. Data on full-genome sequences of SARS-CoV-2 detected in the territory of Primorsky Krai (2020–2023) in the VGARus database
Year | G.Р. Somov Institute of Epidemiology and Microbiology (Vladivostok) | State Research Center of Virology and Biotechnology “Vector" (Novosibirsk) | Institute of Influenza (St.-Petersburg) | Total |
2020 | 173 | 2 | 23 | 198 |
2021 | 90 | 11 | 1 | 102 |
2022 | 105 | 158 | 306 | 569 |
2023 | 185 | 0 | 1 | 186 |
Total | 553 | 171 | 331 | 1055 |
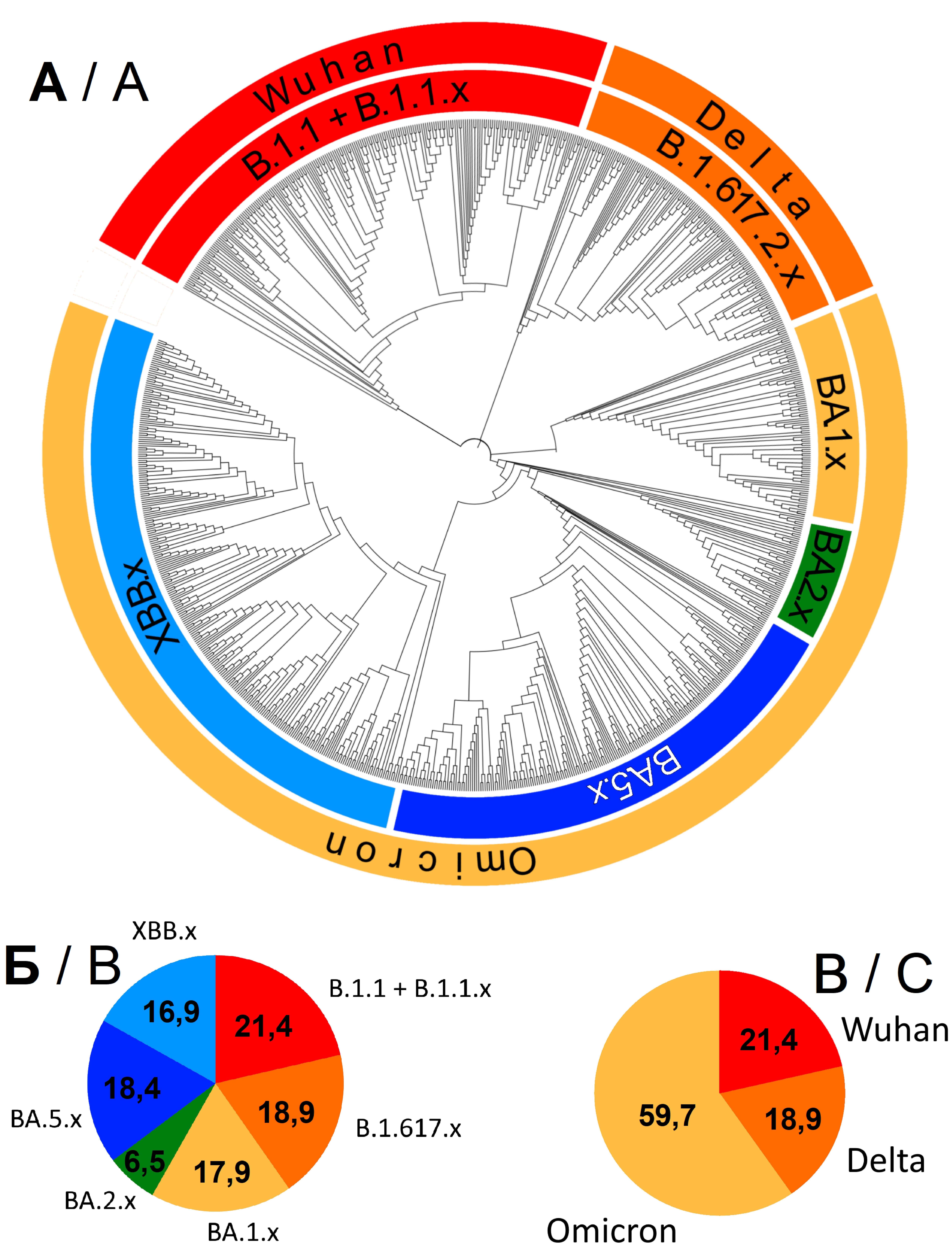
The results of phylogenetic analysis of full-length SARS-CoV-2 genomes from the PK territory (Table 1), shown in Fig. 1, a, indicate differentiation into genetic variants according to the Pango (Fig. 1, b) and WHO (Fig. 1, c) classifications.
Fig. 1. Genotyping of 804 full-genome SARS-CoV-2 nucleotide sequences identified in Primorsky Krai (2020–2023). a — phylogenetic tree; b — distribution according to Pango genetic lines; c — distributions according to WHO genovariants.
The epidemic dynamics of COVID-19 in connection with the change of SARS-CoV-2 genetic variants on the territory of the PK (2020–2023) is presented in Fig. 2 and Table 2: as in Russia as a whole, the rise in COVID-19 incidence during the pandemic period of this disease (11.03.2020–05.05.2023), which began with large megacities [17, 19], had a pronounced wave-like character with 7 epidemic periods.
Fig. 2. Incidence of COVID-19 and genetic diversity of SARS-CoV-2 on the territory of Primorsky Krai (2020–2023).
Table 2. Characteristics of epidemic dynamics of COVID-19 in Primorsky Krai in comparison with a whole Russian Federation (2020–2023)
Period | Territory | Dates of epidemic periods | Incidence (per 100 000 population) | Clinical form | ||||||||||||||
beginning | end | duration, weeks | incidence in the beginning of the period | incidence in the end of the period | maximum | M | m | Me | LQ | UQ | total number of incidences | severe, % | average severity, % | mild and asymptomatic, % | ||||
date | number of a week | date | number of a week | |||||||||||||||
I | PK | 20.04.2020 | 17 | 20.09.2020 | 38 | 22 | 12,83 | 2,65 | 41,8 | 27,6 | 1,7 | 28,2 | 21,1 | 32,6 | 606,6 | 2,1 | 31,5 | 66,4 |
RF | 30.03.2020 | 13 | 30.08.2020 | 36 | 22 | 19,6 | 25,6 | 54,5 | 29,8 | 3,0 | 29,8 | 25,1 | 38,8 | 654,9 | 4,5 | 47,7 | 47,8 | |
II | PK | 21.09.2020 | 39 | 09.05.2021 | 18 | 33 | 28,7 | 9,5 | 91,6 | 54,0 | 5,4 | 54,9 | 24,2 | 87,2 | 1783,4 | 5,8 | 32,2 | 62 |
RF | 31.08.2020 | 36 | 09.05.2021 | 18 | 36 | 28,1 | 41,3 | 154,9 | 73,4 | 6,2 | 60,6 | 42,5 | 101,7 | 2641,9 | 3,1 | 41,6 | 55,3 | |
III | PK | 10.05.2021 | 19 | 03.10.2021 | 39 | 21 | 11,8 | 82,6 | 107,7 | 67,4 | 7,4 | 83,7 | 32,6 | 93,5 | 1415,9 | 3,1 | 35,6 | 61,3 |
RF | 10.05.2021 | 19 | 12.09.2021 | 37 | 18 | 39 | 101,6 | 119,2 | 83,6 | 7,0 | 93,5 | 47,6 | 110,3 | 1504,3 | 2,6 | 41,1 | 56,3 | |
IV | PK | 04.10.2021 | 40 | 09.01.2022 | 1 | 14 | 86,67 | 100,7 | 177,1 | 142,9 | 9,3 | 157,7 | 109,8 | 171,8 | 2001,1 | 3,0 | 36,5 | 60,5 |
RF | 13.09.2021 | 37 | 09.01.2022 | 1 | 17 | 115,7 | 95,5 | 191,8 | 141,8 | 8,3 | 145,5 | 115,7 | 172,8 | 2411,1 | 2,2 | 40,9 | 56,8 | |
V | PK | 10.01.2022 | 2 | 18.07.2022 | 29 | 27 | 104,8 | 8,7 | 581,5 | 144,2 | 35,8 | 56,9 | 11,1 | 149,6 | 3893,5 | 1,3 | 29,8 | 68,9 |
RF | 10.01.2022 | 2 | 18.07.2022 | 29 | 27 | 76,5 | 15,6 | 915,1 | 201,3 | 52,2 | 65,8 | 21,5 | 240,6 | 5435,3 | 0,4 | 33,0 | 66,6 | |
VI | PK | 19.07.2022 | 29 | 09.01.2023 | 2 | 25 | 10,8 | 20,4 | 232,2 | 69,9 | 14,0 | 30,7 | 18,3 | 107,3 | 1747,7 | 0 | 12,1 | 87,9 |
RF | 19.07.2022 | 26 | 09.01.2023 | 2 | 25 | 20,3 | 18,0 | 258 | 91,8 | 16,2 | 44,9 | 29,9 | 164,9 | 2296 | – | – | – | |
VII | PK | 10.01.2023 | 2 | 25.06.2023 | 25 | 23 | 14,3 | 3,04 | 44,4 | 20,1 | 2,5 | 16,1 | 12,7 | 28,7 | 482,2 | 0 | 25,1 | 74,9 |
RF | 10.01.2023 | 2 | 04.06.2023 | 22 | 20 | 15,2 | 8,8 | 64,9 | 36,2 | 3,9 | 34,4 | 21,3 | 54,7 | 767,1 | – | – | – | |
Note. PK — Primorsky Krai; RF — Russian Federation; “–” — no data.
In PK, the first case of COVID-19 was registered on 21.03.2020 (a 30-year-old woman, Russian citizen, living in Vladivostok, who returned on 15.03.2020 from a business trip to Cancun, Mexico), and as of 05.05.2023, 217,196 cases of this disease were documented.
Generally speaking, the dynamics of the epidemic process in the region corresponded to the average in Russia (Fig. 2, Table 2), but with some delay (about 2 weeks) compared to the central regions of the country (similarly to the spread of pandemic influenza A (H1N1 pdm09) [18]). The first rise in COVID-19 incidence in PK was characterized by a slow increase over 13 weeks with a peak at week 29. In Russia, this process developed more rapidly – the peak incidence was registered already in 5 weeks from the beginning of the upsurge with a local maximum in 19 weeks. The slower development of epidemic growth in PK was primarily due to the introduction of strict regime-restrictive measures. During this period in Russia, mainly Wuhan-like variants of the pathogen were isolated from COVID-19 patients: B.1.1 (62.0%), B.1.1.317 (3.3%). In PK, most of the virus genetic variants also belonged to the B.1.1 (26%) and B.1.1.x (B.1.1.317 and B.1.1.277.2) lineages — 74% (Fig. 2, Table 3).
Table 3. Distribution of SARS-CoV-2 genetic variants by COVID-19 pandemic periods on the territory of Primorsky Krai and a whole Russian Federation (2020–2023).
Period | Territory | Number of samples | Genovariants, % | |||||||
Wuhan | Alpha | Delta | Omicron | not found in PK | ||||||
B.1.1 + B.1.1.х | B.1.1.7.x | B.1.617.2.x | BA.1.x | BA.2.x | BA.5.x | XBB.x | ||||
I | PK | 23 | 100,0 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 |
RF | 1701 | 65,3 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 | 34,7 | |
II | PK | 201 | 100,0 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 |
RF | 7417 | 55,0 | 7,2 | 1,6 | 0,0 | 0,0 | 0,0 | 0,0 | 36,2 | |
III | PK | 30 | 3,3 | 0,0 | 96,6 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 |
RF | 10,602 | 5,1 | 2,5 | 89,6 | 0,0 | 0,0 | 0,0 | 0,0 | 2,8 | |
IV | PK | 46 | 2,2 | 0,0 | 97,8 | 0,0 | 0,0 | 0,0 | 0,0 | 0,0 |
RF | 23,315 | 0,1 | 0,0 | 93,5 | 6,1 | 0,0 | 0,0 | 0,0 | 0,3 | |
V | PK | 368 | 0,0 | 0,0 | 33,4 | 48,9 | 17,7 | 0,0 | 0,0 | 0,0 |
RF | 24,416 | 0,1 | 0,0 | 11,4 | 38,9 | 42,5 | 6,2 | 0,0 | 0,9 | |
VI | PK | 201 | 0,0 | 0,0 | 1,0 | 0,0 | 1,0 | 98,0 | 0,0 | 0,0 |
RF | 28,300 | 0,0 | 0,0 | 0,0 | 2,1 | 11,4 | 85,7 | 0,7 | 0,1 | |
VII | PK | 130 | 0,0 | 0,0 | 0,9 | 0,0 | 2,7 | 1,1 | 96,2 | 0,0 |
RF | 4928 | 0,0 | 0,0 | 0,0 | 1,5 | 0,3 | 35,9 | 62,3 | 0,0 | |
Note. PK — Primorsky Krai; RF — Russian Federation.
During wave II, the increase in incidence in both PK and Russia was slower — 11 and 17 weeks, with peaks at weeks 50 and 52, respectively. The proportion of B.1.1 in Russia decreased to 33.7%, while the proportion of B.1.1.317 and B.1.1.523 increased to 21.3%; new genetic variants appeared: Alpha (B.1.1.7) — 7.2% and Delta (B.1.617.2) — 1.6%. In PK, the B.1.1 genetic lineage dominated during this period (68.4%); other gene variants B.1.1.x (primarily B.1.1.317 and B.1.1.277.2) together accounted for 31.6%. In 2021, the periods of rises in incidence were shorter but characterized by a more intensive development of the epidemic process. At that time, III and IV rises of incidence rates were registered in PK with peaks at 27 and 48 weeks, respectively; in Russia, the III rise lasted on average 4 months with the maximum incidence rate at week 29, the IV rise — from September to January 2021. During this period, the process of changing the biological properties of the pathogen was actively underway: the percentage of the Delta genetic variant (B.1.617.2.x) increased to 93–97%, practically displacing all previously circulating lineages.
The fifth rise in incidence in Russia as a whole and in PK was characterized by a sharp increase — the peak of incidence was reached 4 and 5 weeks after the beginning of the rise. The incidence rates during this period were significantly (p < 0.05) higher than in the previous periods. This increase was due to the combined circulation of the outgoing Delta variant (B.1.617.2.x) and the first two lineages of the Omicron variant (BA.1.x and BA.2.x). By the end of Period V, the dominance of the Omicron variant was established worldwide. Omicron BA.5.x sub-lineages, the percentage of which increased up to 90%, caused the VI upsurge of disease incidence in different regions of Russia. At the same time, both in PK and in Russia as a whole, the course of the epidemic process differed from the previous period by a lower intensity and speed of reaching peak indices. During the VII period there was a further decrease in the intensity of the epidemic process. The maximum value of the peak incidence rate on average in Russia was registered in the 8th week, in PK — in the 9th week from the beginning of the massive increase. During this period, the diversity of Omicron sub-variants increased significantly, one of them (XBB) spread rapidly around the world [33]. By the end of January 2023, XBB.x sub-variants prevailed in Russia as a whole as well as in PK, and their percentage in the structure of Omicron sub-variants increased to 62.3 and 92.8%, respectively.
When analyzing the clinical symptoms of COVID-19 in PK, much like in Russia as a whole, infections of mild (68.4 and 54.2%, respectively) and moderate (29.3 and 42.7%) severity prevailed. The percentage of severe forms was 2.3 and 3.1%, respectively. The first wave was characterized by a high proportion of pneumonias (20.9%) and asymptomatic forms (30.3%), the proportion of severe forms was 2.1% (Table 2). As the epidemic process developed in wave II, the highest proportion of severe forms of the disease (5.8%) was observed over the entire observation period, the proportion of non-nosocomial pneumonias was 17.7%. During the subsequent periods, the proportion of non-nosocomial pneumonias gradually decreased from 16.2% in wave III to 1.9% in wave VI. The percentage of severe forms of pneumonia also decreased from 3.1% to 0. It should be noted that the percentage of moderately severe forms of COVID-19 course was distributed unevenly over the periods of the upswing: it increased from I to III period (from 31.5 to 36.5%), decreased to 12.1% in VI period and increased again to 25.1% in VII period.
In epidemic period VII (May 2023), the XBB.x variant, first registered on 31.01.2023 (XBB.1.14 lineage), prevailed in PK, by the end of March — XBB.1.5.24 lineage, a subspecies of variant XBB.1.5 (Kraken), by 05.05.2023 the XBB.1.9.1 (Hyperion) variant became dominant (Fig. 3). It is particularly important to note the high proportion of the XBB.1.5.24 (9% in PK and 6% in Russia as a whole) and XBB.1.9.1 (45 and 51%, respectively) variants, which contrasts sharply with the genetic landscape of SARS-CoV-2 strains in the neighboring countries of South-East Asia (Fig. 4). This pattern may be a consequence of the timely introduction of strict restrictive measures in Russia, including the cessation of transport links with Asia-Pacific countries, while the relative freedom of transport links between Russia and Europe as well as PK and central Russia was maintained throughout the pandemic.
Fig. 3. Genetic diversity of SARS-CoV-2 in Primorsky Krai from January to May 2023.
Fig. 4. Genetic diversity of SARS-CoV-2 in Primorsky Krai, whole Russia, and neighboring countries of East Asia on the eve of the official end of the pandemic on May 05, 2023 (figures in the sectors of pie charts correspond to the percent portion of this genovariant according to GISAID).
Isolation of SARS-CoV-2 strains was successfully performed from 151 samples of RT-qPCR-positive nasopharyngeal washings (isolation efficiency was 151/217 ≈ 69.6%). The obtained viral strains were deposited in the collection of pathogens of the G.P. Somov Research Institute of Epidemiology and Microbiology. Signs of cytopathic effect of the virus in Vero-E6 cell culture were observed from 2–4 days after infection (Fig. 5, a, b). The distribution of the obtained SARS-CoV-2 strains by genetic lineages is presented in Fig. 5, c, and by lg TCID50 values — in Table 4.
Fig. 5. SARS-CoV-2 strains isolated in the sensitive Vero-E6 cell line at the G.P. Somov Institute of Epidemiology and Microbiology of Rospotrebnadzor. a — intact cell line; b — cell line with manifestations of cytopathogenic action of the SARS-CoV-2 strain/Russia_Primorje/FE-R-6932 (Delta AY.122) for 5 days after infection (3 passage); с — the distribution of strains by genovariants (the numbers in the sectors of the pie charts correspond to the percentage of this genovariant).
Table 4. Infectious titers of SARS-CoV-2 strains isolated at the G.P. Somov Institute of Epidemiology and Microbiology (2020–2023)
Genovariant of the strain | Number of strains | Distribution of strains according to the values of the decimal logarithm of the infectious titer, lg IT | M ± m | |||||
3 | 4 | 5 | 6 | |||||
Wuhan | B.1.1 | 49 | 2 | 8 | 11 | 28 | 5,3 ± 0,1 | 5,3 ± 0,1 |
B.1.1.1 | 2 | 0 | 0 | 1 | 1 | 5,5 ± 0,5 | ||
B.1.1.317 | 4 | 0 | 1 | 2 | 1 | 5,0 ± 0,4 | ||
B.1.1.397 | 5 | 0 | 0 | 2 | 3 | 5,6 ± 0,2 | ||
B.1.1.485 | 1 | 0 | 0 | 1 | 0 | 5,0 ± 0,0 | ||
B.1.143 | 1 | 0 | 1 | 0 | 0 | 4,0 ± 0,0 | ||
Delta | AY.121 | 1 | 0 | 0 | 1 | 0 | 5,0 ± 0,0 | 4,3 ± 0,2 |
AY.122 | 36 | 8 | 14 | 9 | 5 | 4,3 ± 0,2 | ||
Omicron | BA.1.x | 20 | 10 | 4 | 6 | 0 | 3,8 ± 0,2 | 3,7 ± 0,1 |
BA.2 | 5 | 2 | 3 | 0 | 0 | 3,6 ± 0,2 | ||
BA.5.2 | 5 | 1 | 3 | 1 | 0 | 4,0 ± 0,3 | ||
XBB.x | 22 | 13 | 8 | 1 | 0 | 3,5 ± 0,1 | ||
The main component plane for different periods of the COVID-19 pandemic in PK and Russia, considered as points in an eight-dimensional feature space, being the components of the SARS-CoV-2 distribution by genetic variants (Table 3), is shown in Fig. 6. The first two main components in Fig. 6, corresponding to the two largest eigenvalues of the feature covariance matrix, include 63.6% of information about the mutual location of objects in the multidimensional feature space. It is known that rotation of the main components by the same angle does not change the values of the eigenvalues of the covariance matrix [32, 38]: in Fig. 6, the rotation of components is performed in such a way as to maximize the variance of factor loadings.
Fig. 6. The plane of the first two main components for various epidemic periods of COVID-19 indicated by Latin numerals (as in Figure 2 and Tables 2–3): the gray dots correspond to the Primorsky Krai, the white ones — to the Russian Federation.
Discussion
In Russia, the COVID-19 pandemic started later than in many countries due to the application of strict regime-limiting and anti-epidemic measures against COVID-19, which were regulated by the Decrees of the President of the Russian Federation7, Orders of the Government of the Russian Federation8 and Resolutions of the Chief State Sanitary Doctor of the Russian Federation9. Measures to prevent the importation and reduce the risk of spread of COVID-19 on the territory of the PK were introduced on the basis of the above mentioned decrees and orders, as well as the Decision of the Governor of the PK10 and the Chief State Sanitary Doctor of the PK11. All the activities carried out at the initial stage made it possible to slow down the spread of COVID-19, increase the volume of COVID-19 tests and the capacity of laboratory bases, prepare medical organizations to provide assistance to patients, which prevented an increase in the number of cases of severe forms of the disease and mortality rate.
The analysis of COVID-19 epidemic process manifestations in the period from April 2020 to June 2023 on the territory of the PK and Russia as a whole revealed 7 epidemiological upsurges of COVID-19 incidence, which differed in clinical and epidemiological manifestations of COVID-19 course depending on the genetic variant of the pathogen (Fig. 1, 2, Tables 2, 3). Comparison of COVID-19 incidence in each period showed that the dynamics of the epidemic process development in PK is similar to the all-Russian development, which is especially clearly demonstrated by Fig. 6. At the same time, mild and moderate clinical forms of the disease prevailed in the structure of clinical forms of the disease throughout the pandemic.
The introduction of restrictive measures in March-April 2020 practically stopped the movement of people between the PK and neighboring countries (China, Republic of Korea, Japan), as a result of which the dynamics of circulation of SARS-CoV-2 virus genetic variants in the region was explained exclusively by intra-Russian migration, therefore, during the I and II periods of the pandemic in the PK (as well as in Russia as a whole) the Wuhan genetic variant was dominant, during the III and IV periods — Delta, and during the V period and subsequent rises — various Omicron genetic variants (Fig. 2). The change of dominant genetic variants is illustrated in Fig. 3.
Since the middle of 2022, a gradual easing of the movement regime with neighboring countries began, which could not but affect the diversity of Omicron genetic variants circulating in PK. Thus, the proportion of XBB.1.9.2 and XBB.1.16 (Arcturus) variants was slightly higher in PK by the end of April — beginning of May 2023 than in Russia as a whole. At the same time, the percentage of the XBB.1.9.2 variant was the highest in the Republic of Korea, and the percentage of the XBB.1.16 variant — in Japan (Fig. 4). These data suggest a certain involvement of neighboring countries of the Asia-Pacific region in shaping the genetic landscape of SARS-CoV-2 in PK at the final stage of the pandemic. The data of phylogenetic analysis of samples of the BN sub-variant, which is rare in Russia (Fig. 5), can also serve as indirect evidence of multiple pathways of virus entry into the region. At the same time, the proportion of registered COVID-19 cases in PK, the etiological factor of which was XBB.1.9.2 (5%) and XBB.1.16 (3%), was comparable to that in the Republic of Korea (XBB.1.9.2 — 7%) and Japan (XBB.1.16 — 12%; Fig. 4), in contrast to the average Russian values (XBB.1.9.2 — 2%, XBB.1.16 — 1%) [34–36]. These data suggest possible participation of the pathogen genetic variants circulating in the neighboring countries of the Asia-Pacific region in the formation of the genetic landscape of SARS-CoV-2 in PK at the final stage of the pandemic.
Phylogenetic analysis of isolates belonging to the BN lineage, which is poorly represented in Russia (about 150 specimens in GISAID) but is characteristic of the Republic of Korea [37], confirms the possibility that new virus variants may have been introduced into PK from this country. According to the Pango website, 37% of isolates of the BN.1.2 lineage, 41% of the BN.1.2.3 lineage, and 25% of the BN.1.3 lineage originate from the PK. Phylogenetic analysis of the most closely related SARS-CoV-2 samples from Russia and the Republic of Korea showed that sample prim000233 (BN.1.2, collection date 14.04.2023) belongs rather to the group of South Korean isolates, while sample prim000314 (BN.1 .2.3, collection date 10.04.2023) belongs to the group of isolates isolated from the central part of Russia (Fig. 7; the low bootstrap support values for some clusters in this figure are explained by the high degree of homology of the analyzed sequences, which is typical of most regional studies of SARS-CoV-2 [38–42]). These results may also serve as indirect evidence of multiple pathways of virus entry into the region.
Fig. 7. Phylogenetic tree of the most homologous viruses of the BN line of SARS-CoV-2. Numbers in VGARus and GISAID (in parentheses) databases are given for the samples from Russia. Numbers in GISAID database are given for the samples from Republic of Korea. Numbers of samples from Primorsky Krai are given in bold.
During the isolation of SARS-CoV-2 strains, a significant decrease in the titers of Delta and Omicron genetic variants was observed, indicating a decrease in their virulence (replication rate and virus accumulation) compared to Wuhan-like variants (Table 4). The obtained results are consistent with the epidemiological manifestations of the infectious process – increased infectivity of SARS-CoV-2 variants and decreased severity of clinical manifestations of infection.
The distribution of SARS-CoV-2 genotypes detected in clinical material (Fig. 1, b, c) shows the predominance of Omicron variant (59,7%), while among isolated strains Wuhan-like variants dominate (41,0%), which is explained by their higher virulence. This confirms the correctness of the choice of a strategy for rapid epidemiological monitoring based on sequencing of the full-length viral genome from primary clinical material (although this does not exclude the need to isolate different variants of the pathogen for virological studies).
Thus, the uniqueness of the SARS-CoV-2 genotypic portrait formation on the territory of the PK during the COVID-19 pandemic lies in the possibility of virus importation both from the European part of Russia and from the countries of East Asia. The latter direction seems to be the most relevant in connection with the strategic "eastern vector" of foreign policy, which is consistently implemented by the Government of the Russian Federation. The continuing possibility of emergence of new, particularly dangerous variants of the virus in neighboring countries makes molecular genetic monitoring for SARS-CoV-2 in the territory of the PK particularly relevant. Retrospective analysis of the epidemic process of COVID-19 in a particular territory is a prerequisite for the formation of models and forecasts of the situation development, allows to identify risk factors associated with the spread and severity of the disease, to assess the effectiveness of measures taken, to increase the reliability of biological safety provision.
1 Rospotrebnadzor reporting form No. 970 "Information on cases of persons with suspected coronavirus infection", report No. 1076 "Summary report on COVID-19 cases in the regions".
2 URL: https://www.rospotrebnadzor.ru/region/korono_virus/epid.php
3 URL: https://стопкоронавирус.рф/
4 URL: https://gisaid.org
5 The number of RT-qPCR-positive samples should not be associated with the number of patients with COVID 19: in 2020, patients were examined an average of 3.6 times, as at that time patients were discharged after a double negative result, which was not achieved the first time; in 2021, discharge started to be performed after a single negative result and later on upon recovery.
6 Order of Rospotrebnadzor of 19.02.2021 № 56 "On improvement of molecular genetic monitoring for strains of the causative agent of a new coronavirus infection".
7 Presidential Decrees: No. 206 of 25.09.2020; No. 239 of 02.04.2020; No. 294 of 02.04.2020.
8 Orders of the Government of the Russian Federation: from 30.01.2020 No. 140-r; from 31.01.2020 No. 154-r; from 03.02.2020 No. 194-r; from 18.02.2020 No. 338-r; from 27.02.2020 No. 447-r; from 27.02. 2020 No. 446-r; of 27.02.2020 No. 448-r; of 16.03.2020 No. 635-r; of 06.03.2020 No. 550-r; of 12.03.2020 No. 597-r; of 14.03.2020 No. 622-r; of 16 March 2020 No. 730-r; of 27.03.2020 No. 763-r.
9 Decisions of the Chief State Sanitary Doctor of the Russian Federation: from 24.01.2020 No. 2; from 31.01.2020 No. 3; from 02.03.2020 No. 5; from 13.03.2020 No. 6; from 18.03.2020 No. 7; from 30.03.2020 No. 9.
10 Resolution of the PK Governor of 18.03.2020 No. 21-pg.
11 Resolution of the Chief State Sanitary Doctor of the PK of 20.04.2020 No. 17.
About the authors
Anna Y. Popova
Federal Service for the Oversight of Consumer Protection and Welfare
Email: uglevas@bk.ru
ORCID iD: 0000-0003-2567-9032
D. Sci. (Med.), Professor, Head, Federal Service for the Oversight of Consumer Protection and Welfare
Russian Federation, MoscowMikhail Y. Shchelkanov
G.Р. Somov Institute of Epidemiology and Microbiology; Far Eastern Federal University
Author for correspondence.
Email: adorob@mail.ru
ORCID iD: 0000-0001-8610-7623
D. Sci. (Biol.), Director, G.Р. Somov Institute of Epidemiology and Microbiology; Head, Department of epidemiology, microbiology and parasitology with the International scientific and educational center for biological safety of Rospotrebnadzor, School of Life Sciences and Biomedicine, Far Eastern Federal University
Russian Federation, Vladivostok; VladivostokNatalia V. Krylova
G.Р. Somov Institute of Epidemiology and Microbiology; Far Eastern Federal University
Email: krylovanatalya@gmail.com
ORCID iD: 0000-0002-9048-6803
D. Sci. (Biol.), leading researcher, Head, Laboratory of respiratory infections, G.Р. Somov Institute of Epidemiology and Microbiology; assistant professor, Department of epidemiology, microbiology and parasitology with the International scientific and educational center for biological safety of Rospotrebnadzor, School of Life Sciences and Biomedicine, Far Eastern Federal University
Russian Federation, Vladivostok; VladivostokAlexey A. Belik
G.Р. Somov Institute of Epidemiology and Microbiology
Email: belik_a_a@mail.ru
ORCID iD: 0000-0002-0303-3188
Cand. Sci. (Biol.), researcher, Laboratory of respiratory infections, G.Р. Somov Institute of Epidemiology and Microbiology
Russian Federation, VladivostokLyubov M. Semeikina
G.Р. Somov Institute of Epidemiology and Microbiology; Center for Hygiene and Epidemiology in Primorsky Krai
Email: lms78@mail.ru
ORCID iD: 0009-0002-1095-7968
postgraduate student, G.Р. Somov Institute of Epidemiology and Microbiology; Deputy Head, Department of epidemiology, Center for Hygiene and Epidemiology in Primorsky Krai
Russian Federation, Vladivostok; VladivostokTatyana S. Zaporozhets
G.Р. Somov Institute of Epidemiology and Microbiology
Email: niiem_vl@mail.ru
ORCID iD: 0000-0002-8879-8496
D. Sci. (Med.), leading researcher, Laboratory of respiratory viral infections, Deputy Director for science, G.Р. Somov Institute of Epidemiology and Microbiology
Russian Federation, VladivostokVyacheslav Y. Smolenskiy
Federal Service for the Oversight of Consumer Protection and Welfare
Email: aidscouncil@gsen.ru
ORCID iD: 0000-0003-1773-4097
Cand. Sci. (Med.), Deputy Head, Federal Service for the Oversight of Consumer Protection and Welfare
Russian Federation, MoscowElena V. Persianova
G.Р. Somov Institute of Epidemiology and Microbiology
Email: helen-pers@yandex.ru
ORCID iD: 0000-0002-5686-8672
Cand. Sci. (Biol.), researcher, Laboratory of respiratory viral infections, G.Р. Somov Institute of Epidemiology and Microbiology
Russian Federation, VladivostokMarina N. Prosyannikova
Center for Hygiene and Epidemiology in Primorsky Krai
Email: marpros67@mail.ru
ORCID iD: 0009-0002-5265-8106
Head, Laboratory of viral and particularly dangerous bacterial infections, Center for Hygiene and Epidemiology in Primorsky Krai
Russian Federation, VladivostokYuriy A. Belov
G.Р. Somov Institute of Epidemiology and Microbiology; Far Eastern Federal University
Email: bornley@yandex.ru
ORCID iD: 0000-0001-8313-5610
junior researcher, Head, Center for molecular diagnostics, G.Р. Somov Institute of Epidemiology and Microbiology; assistant, Department of epidemiology, microbiology and parasitology with the International scientific and educational center for biological safety of Rospotrebnadzor, School of Life Sciences and Biomedicine, Far Eastern Federal University
Russian Federation, Vladivostok; VladivostokOlga V. Iunikhina
G.Р. Somov Institute of Epidemiology and Microbiology; Far Eastern Federal University
Email: olga-iun@inbox.ru
ORCID iD: 0000-0002-6723-582X
Cand. Sci. (Med.), Head, Laboratory of zoonotic infections, G.Р. Somov Institute of Epidemiology and Microbiology; assistant professor, Department of epidemiology, microbiology and parasitology with the International scientific and educational center for biological safety of Rospotrebnadzor, School of Life Sciences and Biomedicine, Far Eastern Federal University
Russian Federation, Vladivostok; VladivostokAnastasiya B. Pott
G.Р. Somov Institute of Epidemiology and Microbiology
Email: adorob@mail.ru
ORCID iD: 0000-0002-1235-4960
researcher, Laboratory of respiratory viral infections, G.Р. Somov Institute of Epidemiology and Microbiology
Russian Federation, VladivostokTatyana F. Khomichuk
Center for Hygiene and Epidemiology in Primorsky Krai
Email: adorob@mail.ru
ORCID iD: 0009-0007-3941-228X
Head, Department of epidemiology, Center for Hygiene and Epidemiology in Primorsky Krai
Russian Federation, VladivostokAnna I. Simakova
Pacific State Medical University
Email: adorob@mail.ru
ORCID iD: 0000-0002-3334-4673
D. Sci. (Med.), Head, Department of infectious diseases, Pacific State Medical University
Russian Federation, VladivostokSvetlana A. Abramova
G.Р. Somov Institute of Epidemiology and Microbiology
Email: adorob@mail.ru
ORCID iD: 0000-0002-2428-3186
junior researcher, Pathology laboratory, G.Р. Somov Institute of Epidemiology and Microbiology
Russian Federation, VladivostokOlga B. Romanova
Center for Hygiene and Epidemiology in Primorsky Krai
Email: adorob@mail.ru
chief physician, Center for Hygiene and Epidemiology in Primorsky Krai
Russian Federation, VladivostokTatyana N. Detkovskaya
Office of the Federal Service for the Oversight of Consumer Protection and Welfare for the Primorsky Krai
Email: adorob@mail.ru
ORCID iD: 0000-0002-7543-0633
Head, Office of the Federal Service for the Oversight of Consumer Protection and Welfare for the Primorsky Krai
Russian Federation, VladivostokSergey P. Kryzhanovskiy
Medical Association of the Far Eastern Branch of the Russian Academy of Sciences
Email: adorob@mail.ru
ORCID iD: 0000-0002-1981-1079
D. Sci. (Med.), Professor, Corresponding member of the Russian Academy of Sciences, Scientific Head, Medical Association, Far Eastern Branch of the Russian Academy of Sciences
Russian Federation, VladivostokNatalia N. Besednova
G.Р. Somov Institute of Epidemiology and Microbiology; Medical Association of the Far Eastern Branch of the Russian Academy of Sciences
Email: adorob@mail.ru
ORCID iD: 0000-0002-2760-9778
д.м.н., профессор, академик РАН, г.н.с. лаб. иммунобиологических препаратов НИИ эпидемиологии и микробиологии им. Г.П. Сомова; главный научный сотрудник научно-исследовательского отдела Медицинского объединения Дальневосточного отделения Российской академии наук
Russian Federation, Vladivostok; VladivostokReferences
- Щелканов М.Ю., Попова А.Ю., Дедков В.Г. и др. История изучения и современная классификация коронавирусов (Nidovirales: Coronaviridae). Инфекция и иммунитет. 2020;10(2):221–46. Shchelkanov M.Yu., Popova A.Yu., Dedkov V.G., et al. History of investigation and current classification of coronaviruses (Nidovirales: Coronaviridae). Russian Journal of Infection and Immunity. 2020;10(2):221–46. DOI: https://doi.org/10.15789/2220-7619-HOI-1412 EDN: https://elibrary.ru/kziwrq
- Никифоров В.В., Колобухина Л.В., Сметанина С.В. и др. Новая коронавирусная инфекция (COVID-19): этиология, эпидемиология, клиника, диагностика, лечение и профилактика. М.;2020. Nikiforov V.V., Kolobukhina L.V., Smetanina S.V., et al. Novel Coronavirus Infection (COVID-19): Etiology, Epidemiology, Clinics, Diagnostics, Treatment, and Prophylaxis. Moscow;2020. EDN: https://elibrary.ru/hgqiyk
- Щелканов М.Ю. Этиология COVID-19. В кн.: COVID-19: от этиологии до вакцинопрофилактики. Руководство для врачей. М.;2023:11–53. Shchelkanov M.Yu. Etiology of COVID-19. In: COVID-19: from Etiology to Vaccine Prevention. Guide for Doctors. Moscow;2023:11–53. DOI: https://doi.org/10.33029/9704-7967-4-COV-2023-1-288 EDN: https://elibrary.ru/gegddl
- Шестопалов А.М., Кононова Ю.В., Гаджиев А.А. и др. Биоразнообразие и эпидемический потенциал коронавирусов (Nidovirales: Coronaviridae) рукокрылых. Юг России: экология, развитие. 2020;15(2):17–34. Shestopalov A.M., Kononova Yu.V., Gadzhiev A.A., et al. Biodiversity and epidemic potential of chiropteran coronaviruses (Nidovirales: Coronaviridae). South of Russia: Ecology, Development. 2020;15(2):17–34 DOI: https://doi.org/10.18470/1992-1098-2020-2-17-34 EDN: https://elibrary.ru/csbxlk
- Щелканов М.Ю., Колобухина Л.В., Бургасова О.А. и др. COVID-19: этиология, клиника, лечение. Инфекция и иммунитет. 2020;10(3):421–45. Shchelkanov M.Yu., Kolobukhina L.V., Burgasova O.A., et al. COVID-19: etiology, clinic, treatment. Russian Journal of Infection and Immunity. 2020;10(3):421–45. DOI: https://doi.org/10.15789/2220-7619-CEC-1473 EDN: https://elibrary.ru/imaadb
- Щелканов М.Ю., Дунаева М.Н., Москвина Т.В. и др. Каталог вирусов рукокрылых (2020). Юг России: экология, развитие. 2020;15(3):6–30. Shchelkanov M.Yu., Dunaeva M.N., Moskvina N.V., et al. Catalog of bat viruses (2020). South of Russia: Ecology, Development. 2020;15(3):6–30. DOI: https://doi.org/10.18470/1992-1098-2020-3-6-30 EDN: https://elibrary.ru/tzqsap
- Щелканов Е.М., Уколов С.С., Дунаева М.Н. и др. Эхолокация рукокрылых (Chiroptera Blumenbach, 1779) как элемент их экологической пластичности. Юг России: экология, развитие. 2020;15(4):6–20. Shchelkanov E.M., Ukolov S.S., Dunaeva M.N., et al. Echolocation of bats (Chiroptera Blumenbach, 1779) as the element of their ecological plasticity. South of Russia: Ecology, Development. 2020;15(4):6–20. DOI: https://doi.org/10.18470/1992-1098-2020-4-6-20 EDN: https://elibrary.ru/mubjcm
- Щелканов М.Ю., Табакаева Т.В., Щелканов Е.М. и др. Паукообразные-эктопаразиты рукокрылых. Владивосток;2022. Shchelkanov M.Yu., Tabakaeva T.V., Shchelkanov E.M., et al. Arachnids-Ectoparasites of Chiropterans. Vladivostok;2022. DOI: https://doi.org/10.24866/7444-5377-0 EDN: https://elibrary.ru/ulhbco
- Щелканов М.Ю., Табакаева Т.В., Щелканов Е.М. и др. Насекомые-эктопаразиты рукокрылых. Владивосток;2022. Shchelkanov M.Yu., Tabakaeva T.V., Shchelkanov E.M., et al. Insects-Ectoparasites of Chiropterans. Vladivostok;2022. DOI: https://doi.org/10.24866/7444-5404-3 EDN: https://elibrary.ru/wfkcuc
- Щелканов М.Ю., Колобухина Л.В., Львов Д.К. Коронавирусы человека (Nidovirales, Coronaviridae): возросший уровень эпидемической опасности. Лечащий врач. 2013;(10):49–54. Shchelkanov M.Yu., Kolobukhina L.V., Lvov D.K. Human coronaviruses (Nidovirales, Coronaviridae): increased level of epidemic threat. The Practitioner. 2013;(10):49–54. EDN: https://elibrary.ru/takhvr
- Щелканов М.Ю., Ананьев В.Ю., Кузнецов В.В., Шуматов В.Б. Ближневосточный респираторный синдром: когда вспыхнет тлеющий очаг? Тихоокеанский медицинский журнал. 2015;(2):94–8. Shchelkanov M.Yu., Ananiev V.Yu., Kuznetsov V.V., Shumatov V.B. Middle East respiratory syndrome: when will smouldering focus outbreak? Pacific Medical Journal. 2015;(2):94–8. EDN: https://elibrary.ru/ulfnff
- Щелканов М.Ю., Ананьев В.Ю., Кузнецов В.В., Шуматов В.Б. Эпидемическая вспышка Ближневосточного респираторного синдрома в Республике Корея (май–июль 2015 г.): причины, динамика, выводы. Тихоокеанский медицинский журнал. 2015;(3):89–93. Shchelkanov M.Yu., Ananiev V.Yu., Kuznetsov V.V., Shumatov V.B. Epidemic outbreak of MERS in the Republic of Korea (May–July, 20015): reasons, dynamics, conclusions. Pacific Medical Journal. 2015;(3):89–93. EDN: https://elibrary.ru/ulhaer
- Kiselev D., Matsvay A., Abramov I., et al. Current trends in diagnostics of viral infections of unknown etiology. Viruses. 2020;12(2):211. DOI: https://doi.org/10.3390/v12020211 EDN: https://elibrary.ru/tvffro
- Акимкин В.Г., Семененко Т.А., Углева С.В. и др. COVID-19 в России: эпидемиология и молекулярно-генетический мониторинг. Вестник Российской академии медицинских наук. 2022;77(4):254–60. Akimkin V.G., Semeneko T.A., Ugleva S.V., et al. COVID-19 in Russia: epidemiology and molecular genetic monitoring. Annals of the Russian Academy of Medical Sciences. 2022;77(4):254–60. DOI: https://doi.org/10.15690/vramn2121 EDN: https://elibrary.ru/dozijs
- Грибова В.В., Окунь Д.Б., Шалфеева Е.А. и др. Облачный сервис для дифференциальной клинической диагностики острых респираторных вирусных заболеваний (в том числе – связанных с особо опасными коронавирусами) методами искусственного интеллекта. Якутский медицинский журнал. 2020;(2):44–7. Gribova V.V., Okun D.B., Shalfeeva E.A., et al. Cloud service for the differential clinical diagnostics of acute respiratory viral diseases (including those associated with highly contagious coronaviruses) with an application of methods of artificial intelligence. Yakut Medical Journal. 2020;(2):44–7. DOI: https://doi.org/10.25789/YMJ.2020.70.13 EDN: https://elibrary.ru/etwrev
- Латыпова М.Ф., Цибин А.Н., Комаров А.Г. и др. Организация геномного надзора за SARS-CoV-2 в структуре департамента здравоохранения города Москвы. Проблемы социальной гигиены, здравоохранения и истории медицины. 2022;30(S):1061–6. Latypova M.F., Tsibin A.N., Komarov A.G., et al. Organization of genomic surveillance for SARS-CoV-2 within the Moscow city health department. Problems of Social Hygiene, Public Health and History of Medicine, Russian Journal. 2022;30(S):1061–6. DOI: https://doi.org/10.32687/0869-866X-2022-30-s1-1061-1066 EDN: https://elibrary.ru/mshhnn
- Акимкин В.Г., Попова А.Ю., Хафизов К.Ф. и др. COVID-19: эволюция пандемии в России. Сообщение II: Динамика циркуляции геновариантов вируса SARS-CoV-2. Журнал микробиологии, эпидемиологии и иммунобиологии. 2022;99(4):381–96. Akimkin V.G., Popova A.Yu., Khafizov K.F., et al. COVID-19: Evolution of the pandemic in Russia. Report II: Dynamics of the circulation of SARS-CoV-2 genetic variants. Journal of Microbiology, Epidemiology and Immunobiology. 2022;99(4):381–96. DOI: https://doi.org/10.36233/0372-9311-295 EDN: https://elibrary.ru/kvulas
- Щелканов М.Ю., Львов Д.Н., Федякина И.Т. и др. Динамика распространения пандемического гриппа А/H1N1 swl на Дальнем Востоке в 2009 г. Вопросы вирусологии. 2010;55(3):10–5. Shchelkanov M.Yu., Lvov D.N., Fedyakina I.T., et al. Trends in the spread of pandemic influenza A(H1N1) swl in the Far East in 2009. Problems in Virology. 2010;55(3):10–5. EDN: https://elibrary.ru/muekiz
- Акимкин В.Г., Попова А.Ю., Плоскирева А.А. и др. COVID-19: эволюция пандемии в России. Сообщение I: Проявления эпидемического процесса COVID-19. Журнал микробиологии, эпидемиологии и иммунобиологии. 2022;99(4):381–96. Akimkin V.G., Popova A.Yu., Ploskireva A.A., et al. COVID-19: Evolution of the pandemic in Russia. Report I: Manifestation of the COVID-19 epidemic process. Journal of Microbiology, Epidemiology and Immunobiology. 2022;99(4):381–96. DOI: https://doi.org/10.36233/0372-9311-276 EDN: https://elibrary.ru/zxgtfd
- Shcheglov B.O., Galkina I.V., Lembikov A.O., et al. Rhinolaryngological simulators based on 3D printing open up new opportunities for professional training. Yakut Medical Journal. 2020;(3):58–60. DOI: https://doi.org/10.25789/YMJ.2020.71.16 EDN: https://elibrary.ru/sxnomh
- Щеглов Б.О., Дунаева М.Н., Баранчугов И.А. и др. Макет дыхательных путей человека. Патент RF 2740727 C1; 2021. Shcheglov B.O., Dunaeva M.N., Baranchugov I.A., et al. Model of human respiratory tract. Patent RF 2740727 C1; 2021. EDN: https://elibrary.ru/hmlszk
- Gonzalez-Recio O., Gutierrez-Rivas M., Peiro-Pastor R., et al. Sequencing of SARS-CoV-2 genome using different nanopore chemistries. Appl. Microbiol. Biotechnol. 2021;105(8):3225–34. DOI: https://doi.org/10.1007/s00253-021-11250-w
- Brejova B., Borsova K., Hodorova V., et al. Nanopore sequencing of SARS-CoV-2: Comparison of short and long PCR-tiling amplicon protocols. PLoS One. 2021;16(10):e0259277. DOI: https://doi.org/10.1371/journal.pone.0259277
- Felsenstein J. Inferring Phylogenies. Sunderland, Massachusetts;2003.
- Крылова Н.В., Иунихина О.В., Федореев С.А. и др. Анти-SARS-CoV-2 активность полифенольного комплекса из Maackia amurensis. Бюллетень экспериментальной биологии и медицины. 2023;176(8):216–9. Krylova N.V., Iunikhina O.V., Fedoreev S.A., et al. Anti-SARS-CoV-2 activity of the polyphenol complex from Maackia amurensis. Bulletin of Experimental Biology and Medicine. 2023;176(8):216–9. DOI: https://doi.org/10.47056/0365-9615-2023-176-8-216-219 EDN: https://elibrary.ru/ivgjae
- Krylova N.V., Kravchenko A.O., Iunikhina O.V., et al. Influence of the structural features of carrageenans from red algae of the Far Eastern seas on their antiviral properties. Marine Drugs. 2022;20(1):60. DOI: https://doi.org/10.3390/md20010060 EDN: https://elibrary.ru/yuukig
- Щелканов М.Ю., Сахурия И.Б., Полякова Е.Б. и др. Повышение качества МТТ-метода с помощью микродозаторных наконечников специальной конструкции. Иммунология. 1998;(4):57–9. Shchelkanov M.Yu., Sakhuria I.B., Polyakova E.B., et al. Improvement of the MTT-based assay by modification of pipette tips. Immunology (Moscow). 1998;(4):57–9. EDN: https://elibrary.ru/mpawbj
- Щелканов М.Ю., Ерёмин В.Ф., Сахурия И.Б. и др. Дегидрогеназная активность инфицированных клеток и биологические свойства различных вариантов ВИЧ-1. Биохимия. 1999;64(4):513–9. Shchelkanov M.Yu., Eremin V.F., Sakhuria I.B., et al. Dehydrogenase activity of infected cells and biological properties of HIV-1 variants. Biochemistry (Moscow). 1999;64(4):431–6. EDN: https://elibrary.ru/lfiulj
- Бейли Н. Математика в биологии и медицине. Пер. с англ. М.;1970. Bailey N. The Mathematical Approach to Biology and Medicine. London-NY-Sydney;1967.
- Урбах В.Ю. Статистический анализ в биологических и медицинских исследованиях. М.;1975. Urbach V.Yu. Statistical Analysis in Biological and Medical Research. Moscow;1975.
- Щелканов М.Ю., Пашкова Т.А., Сахурия И.Б. и др. Анализ биологических характеристик первичных изолятов ВИЧ-1 с помощью метода главных компонент. Вопросы вирусологии. 1998;43(3):117–21. Shchelkanov M.Yu., Pashkova T.A., Sakhuria I.B., et al. Analysis of biological characteristics of primary HIV-1 isolates using the main components method. Problems in Virology. 1998;43(3):117–21. EDN: https://elibrary.ru/mqdzqr
- Щелканов М.Ю., Юдин А.Н., Бурунова В.В. и др. Применение метода главных компонент для анализа эффективности панелей эпитоп-имитирующих пептидов при серотипировании ВИЧ. Иммунология. 1999;20(3):13–8. Shchelka- nov M.Yu., Yudin A.N., Burunova V.V., et al. Application of the basic components method for analysis of effectiveness of epitope-imitating peptides panels in HIV serotyping. Immunology (Moscow). 1999;20(3):13–8. EDN: https://elibrary.ru/yrqiqn
- Tamura T., Ito J., Uriu K., et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat. Commun. 2023;(14):2800. DOI: https://doi.org/10.1038/s41467-023-38435-3
- Yamasoba D., Uriu K., Plianchaisuk A., et al. Virological characteristics of the SARS-CoV-2 omicron XBB.1.16 variant. Lancet Infect. Dis. 2023;23(6):655–6. DOI: https://doi.org/10.1016/s1473-3099(23)00278-5
- WHO. XBB.1.16 Updated Risk Assessment; 05 June 2023. Available at: https://www.who.int/docs/default-source/coronaviruse/05062023xbb.1.16.pdf?sfvrsn=f1845468_3
- Lee D.W., Kim J.M., Park A.K., et al. Genomic epidemiology of SARS-CoV-2 Omicron variants in the Republic of Korea. Sci. Rep. 2022;12(1):22414. DOI: https://doi.org/10.1038/s41598-022-26803-w
- Kim I.H., No J.S., Kim J.A., et al. Genomic epidemiology of SARS-CoV-2 variants in South Korea between January 2020 and February 2023. Virology. 2023;587:109869. DOI: https://doi.org/10.1016/j.virol.2023.109869
- Браверман Э.М., Мучник И.Б. Структурные методы обработки эмпирических данных. М.;1983. Braverman E.M., Muchnik I.B. Structural Methods of Empirical Data Processing. Moscow;1983.
- Basheer A., Zahoor I. Genomic epidemiology of SARS-CoV-2 divulge B.1, B.1.36, and B.1.1.7 as the most dominant lineages in first, second, and third wave of SARS-CoV-2 infections in Pakistan. Microorganisms. 2021;9(12):2609. DOI: https://doi.org/10.3390/microorganisms9122609
- Goletic T., Konjhodzic R., Fejzic N., et al. Phylogenetic pattern of SARS-CoV-2 from COVID-19 patients from Bosnia and Herzegovina: lessons learned to optimize future molecular and epidemiological approaches. Bosn. J. Basic Med. Sci. 2021;21(4):484–7. DOI: https://doi.org/10.17305/bjbms.2020.5381
- Menasria T., Aguilera M. Genomic diversity of SARS-CoV-2 in Algeria and North African Countries: What we know so far and what we expect? Microorganisms. 2022;10(2):467. DOI: https://doi.org/10.3390/microorganisms10020467
- Градобоева Е.А., Тюлько Ж.С., Фадеев А.В. и др. Сравнительный анализ разнообразия линий SARS-CoV-2, циркулирующих в Омской области в 2020–2022 годах. Эпидемиология и вакцинопрофилактика. 2022;21(6):24–33. Gradoboeva E.A., Tyulko J.S., Fadeev A.V., et al. Comparative analysis of the diversity of SARS-CoV-2 lines circulating in the Omsk region in 2020-2022. Epidemiology and Vaccinal Prevention. 2022;21(6):24–33. DOI: https://doi.org/10.31631/2073-3046-2022-6-24-33
Supplementary files