Hepatitis B virus preCore/Core region variability in pregnant women in the Republic of Guinea
- Authors: Ostankova Y.V.1, Balde T.A.2, Boumbaly S.3, Serikova E.N.1, Zueva E.B.1, Reingardt D.E.1, Schemelev A.N.1, Davydenko V.S.1, Anufrieva E.V.1, Esaulenko E.V.1, Totolian A.A.1
-
Affiliations:
- St. Petersburg Pasteur Institute
- Research Institute of Applied Biology
- Centre International de Recherche sur les Infections Tropicales en Guinée
- Issue: Vol 101, No 1 (2024)
- Pages: 61-71
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18522
- DOI: https://doi.org/10.36233/0372-9311-447
- EDN: https://elibrary.ru/waiiez
- ID: 18522
Cite item
Abstract
Introduction. The vertical route of hepatitis B virus (HBV) transmission is a significant problem in African countries, which is characterized by late diagnosis of the disease and high mortality. The high prevalence of hepatocellular carcinoma (HCC) in Africa may be due to variability in the HBV preCore/Core region, mutations in which contribute to disease progression. Molecular genetic characterization of strains circulating among pregnant women may reflect the overall mutational profile of the pathogen in the population.
The objective of this study was to analyze the variability of the HBV preCore/Core region circulating among pregnant women in the Republic of Guinea.
Materials and methods. The study material included 480 plasma samples obtained from HBV-positive pregnant women from the Republic of Guinea. For all samples, the nucleotide sequences of the preCore/Core region of the HBV genome were sequenced and analyzed.
Results. Amino acid variability in the preCore region was determined in 211 (43.96%), and in the Core region in 473 (98.54%) patients. 12 polymorphic sites of the preCore region were identified in which amino acid substitutions occurred, including 8, 2 and 5 positions identified for genotypes E, A and D, respectively. In the Core region, 67 substitution positions were identified, including 46 in samples of genotype E, 23 in HBV genotype A and 26 in genotype D. It was shown that the distribution of substitutions in the preCore and Core regions in HBV genotypes E, A and D differs significantly with a predominance in mutations among HBV genotype E — p < 0.0001. Individual characteristic mutations have been identified for each genotype. The most common clinically significant mutations in the preCore/Core region in the study group were identified, including pc-H5D (27,08%), pc-W28* (35,21%), c-E64D (33,54%), c-L116I/V/G (91,46 %), c-T146N (73,13%). The double mutation A1762T/G1764A in the basal core promoter was shown in 74 samples of HBV genotype E, which accounted for 15.42% of the total group and 16.59% of patients with HBV genotype E.
Conclusion. The frequency of clinically significant preCore/Core mutations among pregnant women in the Republic of Guinea was determined. The data obtained reflect their prevalence in the general population and can be used to predict the progression of chronic HBV among the region's population.
Full Text
Introduction
Hepatitis B virus (HBV), which is capable of causing both acute and chronic liver disease and is the seventh leading cause of death worldwide, remains a serious public health problem despite all measures taken for its elimination. According to various researchers, the number of patients chronically infected with HBV in the world amounts to 360 million people1. More than 75 million of them live in sub-Saharan Africa, where the population prevalence of the HBV surface antigen (HBsAg), the main laboratory diagnostic marker, exceeds 8% and in some regions it could even reach 25% [1]. Natural routes of transmission include sexual (direct sexual contact), vertical (from mother to fetus during or after childbirth), as well as prenatal (transplacental) infection, and transmission due to household contacts, including direct and indirect, including sharing hygiene items and similar methods of contact with an infected person. One of the most significant routes of HBV transmission in Africa is vertical transmission [2]. Without therapeutic intervention, the incidence of mother-to-child transmission exceeds 31% [3]. The particular importance of early infection is due to the fact that when infected before the age of 5 years, the vast majority of cases develop chronic HBV [4]. Furthermore, early HBV infection is one of the most significant risk factors for liver cirrhosis (LC) and hepatocellular carcinoma (HCC) development [5]. Another reason for the huge number of HCC patients and the high mortality rate in Africa is the late diagnosis of the disease and the consequent late referral of patients to health facilities [6]. The most affected country is Gambia, followed by the Republic of Guinea, Liberia and Sierra Leone [7]. This is due to both insufficient coverage with diagnostics and very low public awareness of viral hepatitis, routes of transmission and the consequences of infection. In addition to social factors, viral factors can influence the progression of the disease. Thus, a high viral load and active viral replication as a result of that increases the risk of LC and HCC, but even a viral load of less than 200 IU/ml does not exclude disease progression [8]. The high heterogeneity of the virus, for which 10 genotypes and more than 40 sub-genotypes are currently described, also has an impact on disease progression [9, 10]. For example, HBV subgenotype C2 is characterized by more frequent chronicity and severe disease course, including the development of HCC, than B2, and viruses of genotypes A and B are more sensitive to therapy than D and C [11].
However, the genotypic factor is not the only parameter contributing to disease progression; natural and selective viral mutations also play a major role. With the exception of rare deletions, most mutations in the preCore/Core region are point mutations, mainly associated with decreased HBeAg levels and/or decreased viral load. Moreover, in the Core region, mutations are localized mainly in the immunogenic regions (MHC classes I + II) and thus may influence disease progression. For example, mutations in the preCore region (G1896A) and in the basal core promoter (BCP) — T1753C, A1762T/G1764A, amino acid substitutions in the Core region — F24Y, E64D, E77Q, A80I/T/V, L116I, E180A — are known to be associated with a severe course of liver disease and development of HCC [12]. Molecular genetic variation in viruses may show links to spatial and temporal variation, i.e. evolution over time, spread in geographic regions, at-risk groups, key populations, changes in transmission routes [13]. Therefore, dynamic monitoring of circulating virus variants in population groups reflecting the situation in the population and potentially capable of spreading the pathogen is quite important for predicting the epidemiological situation. Pregnant women are a group which fit that category, as they demonstrate the epidemiological profile of the sexually mature heterosexual population of a particular geographical region under study.
The Republic of Guinea, a country of more than 13.6 million people on the Atlantic coast of West Africa, is one of the poorest countries in the world2, where medicine remains the least publicly funded area. Thus, in 2019, the country's GDP contribution to the health and social work sectors did not exceed 3.2%3. Prevention of vertical transmission of HIV, a key factor in improving maternal and child health, has been significantly reduced, and the average number of visits to antenatal care clinics has decreased [14, 15]. At the same time, the prevalence of HBV in the country is extremely high: unlike HIV, testing for viral hepatitis markers is rarely performed, even during antenatal check-ups of pregnant women, so most patients are not diagnosed with the disease. Since there is no screening for viral hepatitis, there is no prevention of vertical transmission of HBV. The incidence of HBV DNA is 22.36% among conventionally healthy individuals [16], 30.4% among blood donors [17], and 26.5 % among pregnant women in some regions of the country [18, 19]. It was previously shown that almost all HBsAg-positive women who gave birth naturally transmitted the virus to the child [20].
Despite the implementation of HBV vaccination in the country in 2006, the vaccination coverage currently does not exceed 47% of the population, which is due to the lack of a well-established infrastructure for storage and transport of vaccines that require compliance with the cold chain, as well as to the socio-cultural peculiarities of the population, distrust of local residents in medical personnel [21].
There are relatively few data on the molecular genetic features of the virus circulating in the Republic of Guinea, and the characterization of the preCore/Core region is very limited in the literature [22].
The objective of this study was to analyze the variability of preCore/Core region of HBV strains circulating among pregnant women in the Republic of Guinea.
Materials and methods
Blood plasma samples obtained from 480 HBV-infected pregnant women living in the Republic of Guinea were used in this study [19]. Laboratory studies were conducted at the Russian-Guinean Scientific Research Centre for Epidemiology and Prevention of Infectious Diseases, located at the Institute of Applied Biology of Guinea in the prefecture of Kindia. The consent of the Ethical Committee of Guinea was obtained for this stage of work (protocol No. 129/CNERS/16 of 31.08.2015). All subjects gave written informed consent to participate in the experiment.
HBV DNA in blood plasma was determined by real-time PCR with hybridization-fluorescence detection using a method developed at the St. Petersburg Pasteur Research Institute of Epidemiology and Microbiology, which allows detection of HBV DNA at low viral load. The sensitivity of the method is 10 IU/ml when extracted from 100 µl of plasma [23]. Nucleotide sequences of complete HBV genomes were obtained using nested PCR based on overlapping primer pairs followed by Sanger sequencing [22]. The obtained HBV sequences were analyzed to identify mutations in the studied region of the HBV genome relative to the consensus sequence of the Mart-B47 virus (HE974377.1, genotype D) [23] to ensure uniformity in the designation of substitution positions and to identify substitutions characteristic of some genotypes but poorly (or not at all) represented in others. Furthermore, all sequences of genotypes A, D and E were analyzed relative to the reference sequences of the corresponding genotypes (GenBank international database numbers AY128092.1, NC_003977.2 and AB032431.1, respectively). The online database tool Geno2Pheno HBV4, as well as Hepatitis B Virus Phylogenetic Typing Tool5 were used, where AY128092.1, NC_003977.2 and AB032431.1 were also selected as reference strains from the proposed strains.
Statistical data processing was performed using the programs MS Excel, Prizm 5.0 (GraphPad Software Inc.), Statistica 8.0 (StatSoft Inc.). The Clopper-Pearson exact interval was used to assess statistical error. The results are presented with 95% confidence interval (95% CI). To assess the reliability of differences between data obtained in pairwise comparisons, the Fisher’s exact test or the χ2 test with Yates correction was used, depending on the characteristics of the samples. A probability value of p < 0.05 was determined as the threshold for the reliability of differences.
Results
The age of the subjects ranged from 13 to 55 years and averaged 25.8 years. The number of HBsAg-negative cases was 188 (39.17%; 95% CI 34.77-43.69%). HBV genotypes determined on the basis of phylogenetic analysis are presented in Table 1 [19].
Table 1. Hepatitis B virus genotypes circulating among pregnant women in the survey group
Genotype | Quantity | Share, % | 95% CI |
А1 | 8 | 1,67 | 0,72–3,26 |
А3 | 7 | 1,46 | 0,59–2,98 |
D1 | 3 | 0,63 | 0,13–1,82 |
D2 | 5 | 1,04 | 0,34–2,41 |
D3 | 11 | 2,29 | 1,15–4,06 |
E | 446 | 92,9 | 90,24–95,05 |
The nucleotide sequences of the viral genome of each sample were analyzed to study the preCore/Core region variability of HBV in the study group. The amino acid variability of the preCore region was determined in 211 individuals (43.96%; 95% CI 39.46–48.53%) and that of the Core-region in 473 individuals (98.54%; 95% CI 97.02–99.41%). Analysis of the preCore region revealed 12 polymorphic sites in which amino acid substitutions occurred, including 8 positions identified for genotype E, 5 for genotype D, and 2 for genotype A. The occurrence of amino acid substitutions in the preCore region is shown in Fig. 1.
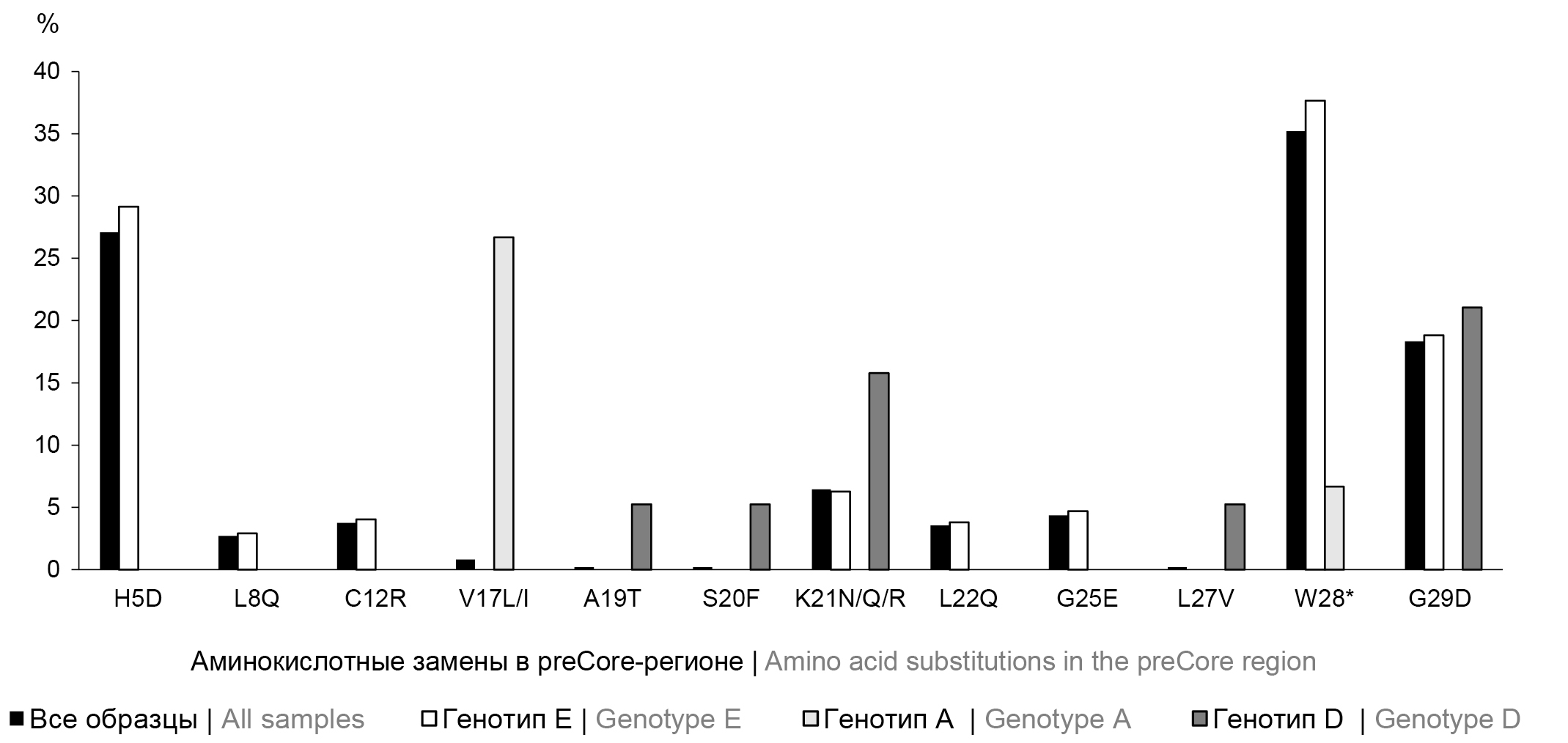
Fig. 1. Frequency of amino acid substitutions occurrence in the preCore region of the hepatitis B virus genome in the examined group and according to genotypes. * — stop codon. Amino acid substitutions are presented relative to the consensus sequence of the Mart-B47 virus (HE974377.1, genotype D) with additional analysis relative to genotype-specific reference sequences AY128092.1 (genotype A), NC_003977.2 (genotype D) and AB032431.1 (genotype E).
During the comparative analysis, the distribution of mutations among HBV genotypes E, A and D was found to be significantly different (p < 0.0001). For genotype E, H5D, L8Q, and C12R substitutions, among others, were identified, which are not represented among isolates of genotypes A and D. In turn, the V17L/I mutation was shown only for HBV genotype A, and substitutions A19T, S20F, L27V were detected only in HBV genotype D. When analyzing the distribution of amino acid substitutions present in at least 2 genotypic groups, significant differences were also identified with a predominance of mutations among HBV genotype E: χ2 = 16.206; df = 8; p = 0.0395.
In the Core region, 67 positions of amino acid substitutions were identified, including 46 for genotype E, 23 for genotype A, and 26 for genotype D. The most common amino acid substitutions occurrence in the Core region is presented in Fig. 2.
Fig. 2. Frequency of the most common amino acid substitutions occurrence in the Core region of the hepatitis B virus genome in the examined group and according to genotypes. Amino acid substitutions are presented relative to the consensus sequence of the Mart-B47 virus (HE974377.1, genotype D) with additional analysis relative to genotype-specific reference sequences — AY128092.1 (genotype A), NC_003977.2 (genotype D) and AB032431.1 (genotype E).
The distribution of Core region mutations among HBV genotypes E, A and D is also significantly different (p < 0.0001). For genotype E, substitutions S21H, V27I, R28K, Y38F, R39G, E46D, P79Q, M93V/I, G94A, F103L, I105T, T114P/V, F122C, W125T, R133T, S141P, L143T, T146N, V149I, R152G, R166P, R181P, Q184R/K/H, were identified, though not represented among HBV genotype A and D samples. Only mutations A11S, A34S, G63V, E77D/Q, L95I, T142I/S, D153G, R159G, S183P were detected among HBV genotype A, and mutations I3L, D4Y, F9I, V13G, L15F, S17L, L19F, D29G, Q57R, L162N, S178T were found among HBV genotype D samples. Evaluation of the distribution of polymorphic variants identified in at least 2 genotypic groups demonstrated significant (p < 0.0001) differences with higher frequency of occurrence and diversity of substitutions among HBV genotype E. Comparative inter-genotype analysis of the most common Core region amino acid substitutions occurrence in the examined group also showed differences (p < 0.0001).
The A1762T/G1764A double mutation was detected in the BCP of 74 HBV genotype E samples, accounting for 15.42% (95% CI 12.3–18.96%) of the total group and 16.59% (95% CI 13.26–20.38%) of patients with HBV genotype E. Information about the most frequent clinically significant BCP and preCore/Core region mutations in the group is presented in Table 2.
Table 2. The most prevalent clinically significant mutations identified in the BCP and preCore/Core region in examined group
HBV genome region | Mutation | Frequency of occurrence in the group (n = 480) | Genotype | Clinical significance of the mutation | ||
n | % | 95% CI | ||||
BCP | A1762T/G1764A | 74 | 15,42 | 12,30–18,96 | E | |
PreCore | H5D | 130 | 27,08 | 23,16–31,30 | E | Associated with severe liver disease in HBsAg-negative HBV genotype E. This mutation could partially explain the high prevalence of HCC in Africa 26] |
PreCore | K21N/Q/R | 31 | 6,46 | 4,43–9,04 | E, D | Presumably associated with disease progression, especially in HBsAg-negative patients [26] |
PreCore | W28* (G1896A) | 169 | 35,21 | 30,93–39,67 | E, A | |
PreCore | G29D (G1899A) | 88 | 18,33 | 14,97–22,09 | E, D | |
Core | E64D | 161 | 33,54 | 29,33–37,96 | E, A, D | Disease progression, liver cirrhosis and HCC, immune escape [24, 25] |
Core | E113Q | 19 | 3,96 | 2,40–6,11 | E, A | Immune escape, selection of specific antibodies, chronic persistence of the virus, disease progression, liver cirrhosis and HCC [24, 25] |
Core | T114P/V | 44 | 9,17 | 6,74–12,11 | E | |
Core | L116I/V/G | 439 | 91,46 | 88,59–93,8 | E, D | |
Core | F122C | 11 | 2,29 | 1,15–4,06 | E | |
Core | W125T | 13 | 2,71 | 1,45–4,59 | E | |
Core | P130T | 29 | 6,04 | 4,08–8,56 | E, D | |
Core | A131P | 67 | 13,96 | 10,98–17,38 | E, A | |
Core | R133T | 11 | 2,29 | 1,15–4,06 | E | |
Core | S141P | 12 | 2,5 | 1,30–4,33 | E | Enhance the formation of HBV cccDNA during intracellular amplification, but impair cccDNA formation during infection [27]. Immune escape, selection of specific antibodies, chronic persistence of the virus, disease progression, liver cirrhosis and HCC [24, 25] |
Core | L143T | 14 | 2,92 | 1,60–4,85 | E | |
Core | T146N | 351 | 73,13 | 68,92–77,04 | E | |
Discussion
The C gene and the Pre-C region of 555 nt and 87 nt, respectively, encode two proteins: HBcAg of 185 amino acids and HBeAg of 150 amino acids, which are the product of alternative translation initiation from two AUG codons and post-translational modification. From the inner AUG codon, the structural polypeptide of the viral capsid Core protein of 21 kDa in size is synthesized. A 24 kDa preCore protein is encoded from the upstream AUG codon. The Core protein plays a key role in the HBV life cycle, and changes in its sequence can serve as potential markers of disease progression [12]. HBcAg serves as a key target for the host immune response, especially attack by CD4 and CD8 cytotoxic T-lymphocytes, whereby nonsynonymous mutations that alter immune epitopes can lead to the formation of escape variants from the immune response, resulting in HBV persistence. Moreover, as substitutions in Core region can lead to simultaneous substitutions in HBeAg, a key immunity regulation protein of the virus, they can have a significant impact on the natural course of chronic HBV [28]. BCP also plays a crucial role in viral replication, contributing to the formation of preCore and pregenomic RNA [29], which means that mutations in this region may also contribute to disease development. Mutations in different regions of the viral genome leading to such changes may be a consequence of both natural evolution of the virus and external influence, since both endogenous and exogenous selective effects lead to modification of the pathogen genome structure due to its high variability, especially during the long-term course of the disease [24].
In the study group, 15 people had HBV genotype A, including 8 cases of genotype A1, which is endemic to Africa and has a long evolutionary history on the continent. It has been previously shown that Africans infected with subgenotype A1 have a 4.5-fold higher risk of developing liver cancer than those infected with other genotypes, with cancer developing at an earlier age [30].
The majority of the cohort studied in the present study consisted of patients with HBV genotype E. This genotype is one of the most common variants in sub-Saharan Africa, including the Republic of Guinea and its neighboring countries. Despite its widespread occurrence, this variant of the virus has a relatively low genome variability compared to others, which may indicate its shorter evolutionary history [31]. At the same time, some researchers have raised concerns about the efficacy of HBV vaccine in genotype E, as breakthrough infections have often been observed in this genotype. Presumably, this may be due to the double mutation A1762T/G1764A in BCP, associated not only with the progression of liver disease, but also with a reduced level of secreted HBeAg, eluding the immune response [32]. Thus, the high incidence of HCC known for HBV genotype E may be related to both the prevalence of genome alterations leading to disease progression and the previously mentioned late detection of infection, with one factor complimenting the other [31]. A high occurrence of the A1762T/G1764A double mutation (15.42%) was shown in our study group. Furthermore, a significant number of mutations in the preCore/Core region of HBV were detected, including a high frequency of the preCore G1896A mutation (35.21%), leading to the appearance of a stop codon (W28*), premature termination of the HBeAg precursor, responsible for more than 90% of cases of defective HBeAg secretion and, consequently, cancellation of antigen expression, which is characteristic of patients infected with HBV genotype E [33]. Note that a similarly high prevalence of the G1896A mutation (47.11%) was previously shown in residents of the Republic of Guinea with HBV genotype E [22]. The preCore region amino acid substitution H5D of HBV genotype E in HBsAg-negative genotype E identified in the present study is associated with severe liver disease. This mutation may also partially explain the high prevalence of HCC in Africa. Two more mutations, K21N/Q/R and G29D in the preCore region, significant for the development of HCC and associated with severe disease in HBsAg-negative individuals, were identified among isolates of genotypes E and D [26]. A significant number of mutations in the Core region at positions 113–143, known to affect antigenicity and particle stability and resulting in immune eluding mutants leading to chronic persistence of the virus, were found in the examined group.
For most of the amino acid substitutions identified in the Core-region, there is no reliable information on their clinical significance; however, HBcAg immune recognition sites are known, including target epitopes for human CD4+ T cells (amino acid positions 1–20, 50–69, 81–105, 117–131, 141–165), cytotoxic T lymphocytes/CD8+ T cells (amino acid positions 18–27, 88–96, 130–140, 141–151), and B–cell epitopes (amino acid positions 74–89, 107–118, 127–138). Mutations in such immunogenic regions of HBcAg are vital for virus persistence, host immune response, and disease progression [25]. Thus, among the detected amino acid substitutions, a number of mutations are of potential clinical significance, contributing to the development of chronic HBV, for example, localized at T cell epitope sites (T12S, E64D, L65V, M66I, T67N/S, A69S, N87G/S, T91N/S, M93V/I, G94A, I97F, F103L, I105T, F122C, W125T, P130T, A131P, S141P, L143T, T146N, V149I, R151Q, R152G, S157I) and B cells (N74V/A, P79Q, A80T, N87G/S, L108I, E113Q, T114P/V, L116I/V/G, P130T, A131P, R133T). Amino acid substitutions in key immune epitopes have the potential to evade the immune response, which in turn may be one of the causes of HBsAg-negative HVB, lead to persistent infection and high variability in all regions of the viral genome [34]. It should be noted that among the listed mutations there are those that are significantly associated with the development of liver cirrhosis and HCC, including those located in B-cell epitopes E77D/Q, A80T and L116I/V/G, as well as in T-cell epitopes E64D and T91N/S. Given the high occurrence of such substitutions, their significant role in the increased prevalence of severe liver diseases in African countries, especially in cases of HBV genotype E, cannot be excluded. The amino acid substitutions of particular interest are those detected in absolutely every genotype E individual examined — E40D, N74V/A, N87G/S, I97F, R181P, E182A/P, which, given their 100% representation, may be characteristic of genotype E in general or associated with the peculiarities of this genetic variant of the virus in the Republic of Guinea, but, on the other hand, are associated with the progression of liver disease.
The study showed greater variability in the preCore/Core region of genotype E compared to A and D, while it is known that HBV genotype E in general is characterized by relative heterogeneity compared to other genotypes. It is apparent that the results of this study contradicting this fact are related to the fact that isolates of genotype E prevailed in the group, while genotypes A and D are represented by single cases.
The occurrence of mutations among HBV genotype E isolates has increased significantly over the past 15 years. In 2009, P. Garmiri et al. demonstrated that in the Republic of Guinea, 26.0% of HBV genotype E cases had 1 or more mutations at positions 1762, 1764 and/or 1896, a premature stop codon associated with the G1896A mutation was observed in 20.8% of sequences, and 5.2% had a triple mutation [35]. In this current study, G1896A presented 1.5 times more frequently (35.21%), and triple mutation was shown in 47 individuals – 9.79% (95% CI 7.28–12.81%) of the total group and 10.54% (95% CI 7.85–13.77%) of those with genotype E. These mutations most likely represent a consequence of natural polymorphism of the virus. However, mutations in positions 1762, 1764, 1896 are among the most common. In Pakistan, A1762T/G1764A was observed in 30% of cases, G1896A — in 38% of cases [36]. In Ethiopia, the occurrence of A1762T/G1764A was 25.9% of cases and G1896A was 25.2% of cases [37]. In Brazil, the occurrence of A1762T/G1764A was 59.3% of cases and G1896A was 84.1% of cases [38]. Furthermore, the occurrence of these mutations is often associated with the virus genotype. For example, the occurrence of A1762T/G1764A in Vietnam was 93.3% for genotype C and 50% for genotype B, and G1896A was 74.2% for genotype B and 2.2% for genotype C [39]. The analysis of 6479 HBV sequences of different genotypes from the international database allowed us to determine the overall frequency of A1762T/G1764A — 28.9%, the representation of this double mutation in relation to genotypes was as follows: A — 26.9%; B — 15.5%; C — 46.1%; D — 21.5%; E — 11%; F — 22.5%; G — 97.5%; H — 3.8%, but it should be noted that the sample volume of genotypes E-H was significantly smaller than the other genotypes [40]. This seems to be the reason for the relatively low occurrence of A1762T/G1764A in genotype E, whereas, for example, in Nigeria, where the study population was predominantly genotype E, A1762T/G1764A was detected in 43% of cases and G1896A in 57% of cases [41].
It should be noted that in our cohort HBsAg-negative and low viral load samples showed more variability throughout the preCore/Core region, which may be related to the inhibition of HBV replication due to mutations in preCore/Core region. Numerous studies have described an association between the frequency of mutations in the preCore/Core region and the progression of liver disease in HBV-infected patients, but the association between amino acid substitutions and clinical severity of disease varies considerably both between different populations and between studies conducted in the same populations. This apparent discrepancy can be explained by various factors, including virus genotype, patient ethnicity, host immune competence, and coinfection with other viruses. Nevertheless, a number of mutations that have been shown to be significantly associated with liver cirrhosis and HCC, as well as affecting HBeAg serological status, may serve as diagnostic and prognostic markers for early detection of liver disease progression in HBV-infected individuals.
Conclusion
The prevalence of clinically significant amino acid substitutions in HBV preCore/Core region in treatment-naïve pregnant women in the Republic of Guinea reflects their occurrence in the population and indirectly explains the reasons for the extremely high incidence of HCC in African countries. A number of such amino acid substitutions, associated with disease progression as shown for genotypes A and D, are characteristic of genotype E. Identification of genotypes and mutations in the virus genome can be used to predict disease progression. The identified variability indicates the need to study the peculiarities of the pathogen and host immune response, including in HBsAg-negative HBV infections. The results of this study demonstrate the necessity to raise awareness of the growing epidemic crisis of HBV in sub-Saharan Africa. This region is at the bottom of the list in terms of access to a range of health care services, including screening, diagnosis and treatment of people infected with the virus. The results of the study can serve as baseline data for estimating national HBV incidence and planning mass HBV immunization in the Republic of Guinea.
1 WHO. Hepatitis B: Fact sheet. 2023. URL: https://www.who.int/ru/news-room/fact-sheets/detail/hepatitis-b
2 Countrymeters. Guinea Population. URL: http://countrymeters.info/en/guinea (дата обращения: 09.03.2023).
3 INS GUINÉE. Annuaire Statistique; 2019. URL: https://www.stat-guinee.org/images/Documents/Publications/INS/annuelles/annuaire/ANNUAIRE_STATISTIQUE_AGRICOLE__2019_INS_FINALISE.pdf (дата обращения: 20.12.2022).
4 URL: https://hbv.geno2pheno.org/
5 URL: https://www.genomedetective.com/app/typingtool/hbv/
About the authors
Yulia V. Ostankova
St. Petersburg Pasteur Institute
Author for correspondence.
Email: shenna1@yandex.ru
ORCID iD: 0000-0003-2270-8897
Cand. Sci. (Biol.), Head, Laboratory of immunology and virology of HIV infection, senior researcher, Laboratory of molecular immunology, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgThierno A.L. Balde
Research Institute of Applied Biology
Email: shenna1@yandex.ru
ORCID iD: 0000-0002-3808-4380
researcher, Research Institute of Applied Biology, Kindia, Republic of Guinea
Guinea, KindiaSanaba Boumbaly
Centre International de Recherche sur les Infections Tropicales en Guinée
Email: shenna1@yandex.ru
ORCID iD: 0000-0002-4506-6033
Cand. Sci. (Biol.), Chief, Graduate school, Research Institute of Applied Biology, Kindia, Republic of Guinea; Director, Centre International de Recherche sur les Infections Tropicales en Guinée
Guinea, NzerecoreElena N. Serikova
St. Petersburg Pasteur Institute
Email: shenna1@yandex.ru
ORCID iD: 0000-0002-0547-3945
junior researcher, Laboratory of immunology and virology of HIV infection, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgElena B. Zueva
St. Petersburg Pasteur Institute
Email: shenna1@yandex.ru
ORCID iD: 0000-0002-0579-110X
Cand. Sci. (Biol.), biologist, Department for diagnosing HIV infection and AIDS-related diseases, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgDiana E. Reingardt
St. Petersburg Pasteur Institute
Email: shenna1@yandex.ru
ORCID iD: 0000-0002-0931-102X
doctor of clinical laboratory diagnostics, Department for diagnosing HIV infection and AIDS-related diseases, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgAlexandr N. Schemelev
St. Petersburg Pasteur Institute
Email: shenna1@yandex.ru
ORCID iD: 0000-0002-3139-3674
junior researcher, Laboratory of immunology and virology of HIV Infection, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgVladimir S. Davydenko
St. Petersburg Pasteur Institute
Email: shenna1@yandex.ru
ORCID iD: 0000-0003-0078-9681
junior researcher, Laboratory of immunology and virology of HIV infection, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgEkaterina V. Anufrieva
St. Petersburg Pasteur Institute
Email: shenna1@yandex.ru
ORCID iD: 0009-0002-1882-529X
junior researcher, Laboratory of immunology and virology of HIV infection, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgElena V. Esaulenko
St. Petersburg Pasteur Institute
Email: shenna1@yandex.ru
ORCID iD: 0000-0003-3669-1993
D. Sci. (Med.), Professor, Head, Laboratory of viral hepatitis, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgAreg A. Totolian
St. Petersburg Pasteur Institute
Email: shenna1@yandex.ru
ORCID iD: 0000-0003-4571-8799
D. Sci. (Med.), Professor, Academician of RAS, Head, Laboratory of molecular immunology, Director, St. Petersburg Pasteur Institute
Russian Federation, St. PetersburgReferences
- Schweitzer A., Horn J., Mikolajczyk R.T., et al. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386(10003):1546–55. DOI: https://doi.org/10.1016/S0140-6736(15)61412-X
- Kean E., Funk A.l., Shimakawa Y. Systematic review with meta-analysis: the risk of mother to child transmission of HBV infection in Sub-Saharan Africa. Aliment. Pharmacol. Ther. 2016;44(10):1005–17. DOI: https://doi.org/10.1111/apt.13795
- Yao N., Fu S., Wu Y., et al. Incidence of mother-to-child transmission of hepatitis B in relation to maternal peripartum antiviral prophylaxis: A systematic review and meta-analysis. Acta Obstet. Gynecol. Scand. 2022;101(11):1197–206. DOI: https://doi.org/10.1111/aogs.14448
- Shimakawa Y., Lemoine M., Njai H.F., et al. Natural history of chronic HBV infection in West Africa: a longitudinal population-based study from The Gambia. Gut. 2016;65(12):2007–16. DOI: https://doi.org/10.1136/gutjnl-2015-309892
- Wang C.C., Cheng P.N., Kao J.H. Systematic review: chronic viral hepatitis and metabolic derangement. Aliment. Pharmacol. Ther. 2020;51(2):216–30. DOI: https://doi.org/10.1111/apt.15575
- Kew M.C., Welschinger R., Viana R. Occult hepatitis B virus infection in Southern African blacks with hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2008;23(9):1426–30. DOI: https://doi.org/10.1111/j.1440-1746.2008.05481.x
- Ladep N.G., Lesi O.A., Mark P., et al. Problem of hepatocellular carcinoma in West Africa. World J. Hepatol. 2014;6(11):783–92. DOI: https://doi.org/10.4254/wjh.v6.i11.783
- Mulrooney-Cousins P.M., Michalak T.I. Persistent occult hepatitis B virus infection: experimental findings and clinical implications. World J. Gastroenterol. 2007;13(43):5682–6. DOI: https://doi.org/10.3748/wjg.v13.i43.5682
- Lin Y.Y., Liu C., Chien W.H., et al. New insights into the evolutionary rate of hepatitis B virus at different biological scales. J. Virol. 2015;89(7):3512–22. DOI: https://doi.org/10.1128/JVI.03131-14
- Lin C.L., Kao J.H. Hepatitis B virus genotypes and variants. Cold Spring Harb. Perspect. Med. 2015;5(5):a021436. DOI: https://doi.org/10.1101/cshperspect.a021436
- Cao G.W. Clinical relevance and public health significance of hepatitis B virus genomic variations. World J. Gastroenterol. 2009;5(46):5761–9. DOI: https://doi.org/10.3748/wjg.15.5761
- Yll M., Cortese M.F., Guerrero-Murillo M., et al. Conservation and variability of hepatitis B core at different chronic hepatitis stages. World J. Gastroenterol. 2020;26(20):2584–98. DOI: https://doi.org/10.3748/wjg.v26.i20.2584
- Mixson-Hayden T., Lee D., Ganova-Raeva L., et al. Hepatitis B virus and hepatitis C virus infections in United States-bound refugees from Asia and Africa. Am. J. Trop. Med. Hyg. 2014;90(6):1014–20. DOI: https://doi.org/10.4269/ajtmh.14-0068
- Camara B.S., Delamou A., Diro E., et al. Effect of the 2014/2015 Ebola outbreak on reproductive health services in a rural district of Guinea: an ecological study. Trans. R. Soc. Trop. Med. Hyg. 2017;111(1):22–9. DOI: https://doi.org/10.1093/trstmh/trx009
- Leno N.N., Delamou A., Koita Y., et al. Ebola virus disease outbreak in Guinea: what effects on prevention of mother-to-child transmission of HIV services? Reprod. Health. 2018; 15(1):60. DOI: https://doi.org/10.1186/s12978-018-0502-y
- Бумбали С., Серикова Е.Н., Семенов А.В. и др. Значимость лабораторной диагностики парентеральных вирусных гепатитов в Гвинейской Республике. Журнал микробиологии, эпидемиологии и иммунобиологии. 2021;98(4):440–9. Bumbali S., Serikova E.N., Semenov A.V., et al. Significance of parenteral viral hepatitis laboratory diagnostics in the Republic of Guinea. Journal of Microbiology, Epidemiology and Immunobiology. 2021;98(4):440–9. DOI: https://doi.org/10.36233/0372-9311-116 EDN: https://elibrary.ru/wdbxjj
- Бумбали С., Балде T.Л., Семенов А.В. и др. Распространенность маркеров вирусного гепатита В среди доноров крови в Гвинейской Республике. Вопросы вирусологии. 2022;67(1):59–68. Bumbali S., Balde T.L., Semenov A.V., et al. Prevalence of viral hepatitis B markers among blood donors in the Republic of Guinea. Problems of Virology. DOI: https://doi.org/10.36233/0507-4088-92 EDN: https://elibrary.ru/zybhjz
- Балде T.A.Л., Бумбали С., Серикова Е.Н. и др. Сравнительный анализ вертикального риска передачи некоторых гемоконтактных инфекций в Гвинейской Республике. Проблемы особо опасных инфекций. 2021;(1):87–94. Balde T.A.L., Bumbali S., Serikova E.N., et al. Comparative analysis of the vertical risk of transmission of some blood-borne infections in the republic of Guinea. Problems of Particularly Dangerous Infections. DOI: https://doi.org/10.21055/0370-1069-2021-1-87-94 EDN: https://elibrary.ru/upnyfx
- Балде T.A.Л., Останкова Ю.В., Бумбали С. и др. Частота встречаемости мутаций лекарственной устойчивости и ускользания от иммунного ответа в геноме вируса гепатита В, выявленного у беременных в Гвинейской Республике. Вопросы вирусологии. 2023;68(3):228–41. Balde T.A.L., Ostankova Yu.V., Bumbali S., et al. Frequency of drug resistance and immune escape mutations in the hepatitis B virus genome detected in pregnant women in the Republic of Guinea. Problems of Virology. DOI: https://doi.org/10.36233/0507-4088-175 EDN: https://elibrary.ru/scvbmy
- Kaba D., Bangoura M.A., Sylla M.M., et al. Prevalence and factors associated with hepatitis B in a cohort of HIV-infected children in the Pediatric Department at Donka National Hospital, Guinea. Pan. Afr. Med. J. 2019;34:182. DOI: https://doi.org/10.11604/pamj.2019.34.182.16275
- Попова А.Ю., Кутырев В.В., Тотолян А.А., ред. Гепатит B в странах Западной Африки: эпидемиология, диагностика, профилактика. СПб.; 2021. Popov A.Y., Kutyrev V.V., Totolyan A.A., ed. Hepatitis B in West African Countries: Epidemiology, Diagnosis, Prevention. St. Petersburg; 2021. EDN: https://elibrary.ru/lpjvsd
- Бумбали С., Серикова Е.Н., Балде Т.Л. и др. Аминокислотные замены в регионах CORE и HBsAg вируса гепатита В при моноинфекции и ВГВ/ВИЧ-коинфекции в Гвинейской республике. ВИЧ-инфекция и иммуносупрессии. 2021;13(3):96–107. Boumbaly S., Serikova E.N., Balde T.A.L., et al. Amino acid substitutions in core and HBsAg regions of hepatitis B virus in patients with monoinfection and HBV/HIV-coinfection in the Republic of Guinea. HIV Infection and Immunosuppressive Disorders. 2021;13(3):96–107. DOI: http://doi.org/10.22328/2077-9828-2021-13-3-122-133 EDN: https://elibrary.ru/vnjjas
- Останкова Ю.В., Серикова Е.Н., Семенов А.В., Тотолян АА. Метод выявления в биологическом материале ДНК вируса гепатита В при низкой вирусной нагрузке на основе гнездовой ПЦР с детекцией по трем вирусным мишеням в режиме реального времени. Клиническая лабораторная диагностика. 2022;67(9):530–7. Ostankova Yu.V., Serikova E.N., Semenov A.V., Totolyan A.A. Method for hepatitis b virus DNA detecting in biological material at low viral load based on nested PCR with detection on three viral targets in real-time mode. Clinical Molecular Studies. 2022;67(9):530–7. DOI: https://doi.org/10.51620/0869-2084-2022-67-9-530-537
- Brichler S., Lagathu G., Chekaraou M.A., et al. African, Amerindian and European hepatitis B virus strains circulate on the Caribbean Island of Martinique. J. Gen. Virol. 2013;94 (Pt. 10):2318–29. DOI: https://doi.org/10.1099/vir.0.055459-0
- Kumar R. Review on hepatitis B virus precore/core promoter mutations and their correlation with genotypes and liver disease severity. World J. Hepatol. 2022;14(4):708–18. DOI: https://doi.org/10.4254/wjh.v14.i4.708
- Al-Qahtani A.A., Al-Anazi M.R., Nazir N., et al. The correlation between hepatitis B virus precore/core mutations and the progression of severe liver disease. Front. Cell Infect. Microbiol. 2018;8:355. DOI: https://doi.org/10.3389/fcimb.2018.00355
- Mendenhall M.A., Hong X., Hu J. Hepatitis B virus capsid: the core in productive entry and covalently closed circular DNA formation. Viruses. 2023;15(3):642. DOI: https://doi.org/10.3390/v15030642
- Kim H., Lee S.A., Do S.Y., Kim B.J. Precore/core region mutations of hepatitis B virus related to clinical severity. World J. Gastroenterol. 2016;22(17):4287–96. DOI: https://doi.org/10.3748/wjg.v22.i17.4287
- Quarleri J. Core promoter: a critical region where the hepatitis B virus makes decisions. World J. Gastroenterol. 2014;20:425–35. DOI: https://doi.org/10.3748/wjg.v20.i2.425
- Kew M.C., Kramvis A., Yu M.C., et al. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-Saharan Africans. J. Med. Virol. 2005;75(4):513–21. DOI: https://doi.org/10.1002/jmv.20311
- Ingasia L.A.O., Kostaki E.G., Paraskevis D., Kramvis A. Global and regional dispersal patterns of hepatitis B virus genotype E from and in Africa: A full-genome molecular analysis. PLoS One. 2020;15(10):e0240375. DOI: https://doi.org/10.1371/journal.pone.0240375
- Malagnino V., Salpini R., Maffongelli G., et al. High rates of chronic HBV genotype E infection in a group of migrants in Italy from West Africa: Virological characteristics associated with poor immune clearance. PLoS One. 2018;13(3):e0195045. DOI: https://doi.org/10.1371/journal.pone.0195045
- Bannister E.G., Yuen L., Littlejohn M., et al. Molecular characterization of hepatitis B virus (HBV) in African children living in Australia identifies genotypes and variants associated with poor clinical outcome. J. Gen. Virol. 2018;99(8):1103–14. DOI: https://doi.org/10.1099/jgv.0.001086
- Sanaei N., Hashemi S.M.A., Dehno S.Z.S., et al. Precore/core mutations of hepatitis B virus genotype D arising in different states of infection. Clin. Exp. Hepatol. 2022;8(1):21–8. DOI: https://doi.org/10.5114/ceh.2022.114253
- Garmiri P., Loua A., Haba N., et al. Deletions and recombinations in the core region of hepatitis B virus genotype E strains from asymptomatic blood donors in Guinea, west Africa. J. Gen. Virol. 2009;90(10):2442–51. DOI: https://doi.org/10.1099/vir.0.012013-0
- Ahmad I., Ahmad K. Molecular characterization of hepatitis B virus basal core promoter and precore region of isolates from chronic hepatitis B patients. J. Pak. Med. Assoc. 2021;71(6): 1575–82. DOI: https://doi.org/10.47391/JPMA.1254
- Belyhun Y., Liebert U.G., Maier M. Analysis of HBV basal core promoter/precore gene variability in patients with HBV drug resistance and HIV co-infection in Northwest Ethiopia. PLoS One. 2018;13(2):e0191970. DOI: https://doi.org/10.1371/journal.pone.0191970
- Chachá S.G.F., Gomes-Gouvêa M.S., Malta F.M., et al. Basal core promoter and precore mutations among hepatitis B virus circulating in Brazil and its association with severe forms of hepatic diseases. Mem. Inst. Oswaldo Cruz. 2017;112(9): 626–31. DOI: https://doi.org/10.1590/0074-02760160540
- Ho P.T., Balzanelli M.G., Distratis P., et al. Characteristics of hepatitis B virus genotype and sub-genotype in hepatocellular cancer patients in Vietnam. Diagnostics (Basel). 2022;12(10): 2393. DOI: https://doi.org/10.3390/diagnostics12102393
- Araujo N.M., Teles S.A., Spitz N. Comprehensive analysis of clinically significant hepatitis B virus mutations in relation to genotype, subgenotype and geographic region. Front. Microbiol. 2020;11:616023. DOI: https://doi.org/10.3389/fmicb.2020.616023
- Grant J., Agbaji O., Kramvis A., et al. Hepatitis B virus sequencing and liver fibrosis evaluation in HIV/HBV co-infected Nigerians. Trop. Med. Int. Health. 2017;22(6):744–54. DOI: https://doi.org/10.1111/tmi.12873
Supplementary files