Characteristics of antibiotic resistance of non-typhoidal Salmonella circulating in the Russian Federation in the period from 2019 to 2022
- Authors: Pavlova A.S.1, Kuleshov K.V.1, Krutova N.E.1, Guseva A.N.1, Podkolzin A.T.1
-
Affiliations:
- Central Research Institute of Epidemiology
- Issue: Vol 100, No 5 (2023)
- Pages: 287-301
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18475
- DOI: https://doi.org/10.36233/0372-9311-451
- EDN: https://elibrary.ru/tmxvam
- ID: 18475
Cite item
Abstract
Introduction. Non-typhoidal Salmonella make a significant contribution to the incidence of enteric infections and are characterized by an increasing proportion of strains resistant to antimicrobial agents (AMA), including the first choice antibiotics (cephalosporins III and fluoroquinolones).
The purpose of the study is to assess the phenotypic resistance of Salmonella to various classes of AMAs and determine the relationship between the phenotypic resistance, serotype, source of isolation and nature of incidence.
Materials and methods. We studied 752 representative strains of Salmonella of 2494 strains isolated from various sources (clinical samples, food products, environment) received from 59 regions of Russia in the period from 2019 to 2022. The phenotypic resistance to 22 antibiotics of 11 CLSI classes of AMAs was assessed by broth microdilution method (minimum inhibitory concentration). The diversity of resistance profiles of Salmonella serotypes was compared using the Shannon index.
Results. The dominant position in terms of isolation frequency is occupied by the serotypes Salmonella Enteritidis, S. Infantis, S. Muenchen, S. Typhimurium, S. Bovismorbificans, which accounted for 64.4% of the studied strains. 543 (72.2%) strains showed resistance to at least one of the tested antibiotics; 193 (25.7%) strains were characterized by multidrug resistance phenotype (MDR). Resistance to AMA classes was characterized by the following distribution: quinolones (61.3%), tetracyclines (28.1%), penicillins (19.1%), β-lactam combination agents (18.6%), folate pathway antagonists (16, 5%), phenicols (10.1%), aminoglycosides (5.6%), cephems (4.7%), monobactams (4.4%), lipopeptides (3.9%). No penem-resistant strains have been identified. The features of Salmonella resistance by AMA classes are shown to depend on the sources of isolation, the Salmonella serotype and the nature of the incidence (outbreak and sporadic).
Conclusions. Monitoring of phenotypic antibiotic resistance is an important tool for epidemiological surveillance in order to prevent the spread of bacterial resistance to AMAs.
Full Text
Introduction
Non-typhoidal strains of Salmonella enterica subsp. enterica (hereafter referred to as non-typhoidal Salmonella) make a significant contribution to the incidence of enteric infections in the world [1, 2]. These strains of Salmonella is responsible for 99% of cases of salmonellosis in humans and animals and remains relevant in the formation of outbreak incidence, ranking 3rd (after acute intestinal infection of viral etiology) by the number outbreaks with the fecal-oral chain of transmission of the infection1. Despite the downward trend in the incidence in recent years, salmonellosis is still the most frequently reported foodborne zoonosis in Russia. According to the dynamics of incidence in Russia from 2019 to 2021, the incidence rate decreased 1.8 times and amounted to 13.61 per 100 thousand population, while in 2022 it was 17.1 per 100 thousand population1.
Uncomplicated salmonellosis, as a rule, does not require antibacterial therapy, but it is indicated for the treatment of invasive and severe forms, as well as patients of high-risk groups2 (infants, elderly people and patients with weakened immune systems) [3, 4]. For a long time, the commonly used antimicrobial agents (AMAs) were chloramphenicol, ampicillin, and trimethoprim-sulfamethoxazole [5, 6], but in recent years, resistance to these AMAs has increased significantly [7].
Currently, the recommended the first choice antibiotics in the treatment of severe and generalized forms of salmonellosis are third-generation cephalosporins and fluoroquinolones3,4. These AMAs are included in the list of mandatory monitoring for Salmonella according to the guidelines of the World Health Organization. The percentage of ciprofloxacin-resistant Salmonella strains isolated from humans increased from 12.0% in 2017 to 19.7% in 2020, according to the latest report from the Global Antimicrobial Resistance and Use Surveillance System (GLASS).5 At the same time, the percentage of detection of extended-spectrum β-lactamase-producing strains, although remaining at a low level, tends to increase [8].
Monitoring of circulating strains and determination of their AMA resistance profiles are necessary to better understand the epidemiological situation, develop national strategies to eradicate resistant strains, and develop preventive measures to avoid the formation of reservoirs of infection. At the same time, the organization of monitoring should be based on the One Health principle, which engage close interaction and coordination of different areas. In particular, the organization of monitoring of the phenotypic and molecular genetic characteristics of Salmonella both in the human population and in different branches of agriculture, which is especially important to ensure food safety [9].
Comparative analysis of data on antibiotic bacterial resistance of Salmonella obtained in Russia in different years involves certain difficulties. One of the main reasons is continuously changing interpretation criteria and methodological approaches to testing for antibiotic bacterial resistance. These changes are occurring around the world, making it difficult to conduct an objective analysis [10]. According to previous studies, the percentage of non-typhoidal Salmonella strains resistant to various AMA classes varied significantly across different regions of Russia in different years. Resistance to aminoglycosides was shown by 1–2% to 53.1% of strains, to quinolones — from 0.0% to 59.2%, to penicillins — from 26.4% to 42.0% [11–14]. Another important factor is the limited number of studies in Russia reflecting a comprehensive analysis of the phenotypic resistance of non-typhoidal Salmonella strains in terms of the diversity of sources and geography of isolation, as well as the serotype diversity.
The purpose of this study was to assess the phenotypic resistance of Salmonella to various AMA classes and to determine the relationship between the phenotypic resistance and such parameters as serotype, isolation source and the nature of the incidence (outbreak and sporadic) at which the strains were isolated.
Materials and Methods
Selection of isolates and microbiological studies
In 2019–2022, 2494 strains of non-typhoidal Salmonella isolated in 59 regions of Russia from various sources were studied. Among them, 1355 isolates were obtained during the investigation of 113 cases of outbreak incidence of salmonellosis from people and suspected infection transmission factors (food and environment), 1139 isolates were obtained in single cases of salmonellosis from food and the environment.The study was conducted with the informed consent of the patients. The research protocol was approved by the Ethics Committee of the Central Research Institute of Epidemiology (protocol No. 83, June 26, 2018).
The studied strains were preliminary cultured to obtain individual colonies on MacConkey medium (CONDA Pronadisa). The species identity was established using biochemical identification systems API® 20E (bioMérieux).
Serological identification
Serological identification of Salmonella was carried out using standardized methods according to the Kaufman–White classification scheme with diagnostic polyclonal sera (PETSAL) and monoclonal sera (Sifin).
Genetic typing
Genetic typing was carried out according to the international standardized protocol using restriction enzymes XbaI and BlnI [15].
Determination of phenotypic resistance
Among all analyzed strains (n = 2494), 1522 isolates (61.0%; 95% confidence interval (CI) 59.1–62.9) were determined to be susceptible to AMAs (729 strains from loci of outbreak incidence and 793 from sporadic cases of detecting Salmonella) by broth microdilution method with determining the minimum inhibitory concentration of the antibiotic on plates G-I and G-II Mikrolatest®SensiLaTest MIC (Erba Lachema). Escherichia coli strain (ATCC 25922) was used as a control strain. The spectrum of AMAs by class, according to the Clinical and Laboratory Standards Institute (CLSI) classification6, included:
- penicillins: ampicillin, piperacillin;
- β-lactam combination agents: ampicillin/sulbactam, piperacillin/tazobactam;
- cephems: cefazolin, cefuroxime, cefotaxime, ceftazidime, cefepime;
- monobactams: aztreonam;
- penems: meropenem, ertapenem;
- aminoglycosides: gentamicin, netilmicin, amikacin, tobramycin;
- lipopeptides: colistin;
- folate pathway antagonists: trimethoprim/sulfamethoxazole;
- quinolones: ciprofloxacin;
- phenicols: chloramphenicol;
- tetracyclines: tetracycline, tigecycline.
The results of susceptibility tests were interpreted in accordance with the current European Committee on Antibiotic Susceptibility Testing criteria (EUCAST v. 13.0). If a strain showed resistance to at least one antibiotic of a certain class, then the strain was considered resistant to this class.
Due to the lack of interpretation of the minimum inhibitory concentrations of cefazolin, cefuroxime, netilmicin, and tigecycline for Salmonella spp., data on these AMAs were not taken into account in the study. The interpretation for tetracycline and chloramphenicol was assessed using epidemiological cut-off values (ECOFF). In addition, following the World Health Organization recommendations, the ECOFF value of >8 µg/ml was established for colistin when testing Salmonella strains belonging to the serotypes S. Enteritidis and S. Dublin7.
Strategy for identifying of representative strains and assessing the diversity of resistance profiles
Representative strains were those strains that were characterized by a unique combination of such characteristics as serotype, isolation date, isolation site, isolation source, AMA resistance profile, and genetic subtype. Each event of outbreak incidence, regardless of the isolation source, was represented by one strain or several strains, if strains differing in one or more characteristics were identified within the outbreak. If there was no genetic subtype for a strain, it was not taken into account in the spectrum of characteristics.
To analyze the diversity of resistance profiles of Salmonella spp., we used the Shannon diversity index (H). The significance of the difference in Shannon index values (p-level of significance) between two groups of strains, each of which was characterized by a set of resistance profiles with certain frequencies of occurrence, was assessed using modified parametric Student’s t-test — Hutcheson’s t-test [16].
The 95% CI for the proportion of detected strains according to various criteria was calculated using the BinomCI function according to the Wilson method using the DescTools library package of the R software.
Results
Patterns and characteristics of resistance of Salmonella circulating in Russia
Among 1522 isolates tested for antibiotic susceptibility in 2019–2022, 752 representative strains belonging to 69 serotypes were selected. Of these, 153 strains belonged to loci of outbreak incidence and included Salmonella strains isolated both from people (n = 121) and from suspected sources (n = 32), and 599 strains were from sporadic cases of salmonellosis (Table 1). Sporadic strains were divided into three groups according to isolation sources: sporadic incidence — strains isolated from humans (n = 270), from food (n = 239) and the enviromental samples — from various water bodies and wastewater (n = 90).
Table 1. Distribution of S. enterica serotypes in outbreaks and in sporadic cases of Salmonella isolation
Serotype (number of outbreaks) | Number of strains | Percentage of strains, % | 95% CI |
Outbreak cases | |||
S. Enteritidis (103) | 138 | 90,2 | 84,4–93,9 |
S. Abony (2) | 5 | 3,3 | 1,4–7,4 |
S. Typhimurium (2) | 3 | 1,9 | 0,6–5,6 |
S. Muenchen (2) | 2 | 1,3 | 0,3–4,6 |
S. Bareilly (1) | 2 | 1,3 | 0,3–4,6 |
S. Infantis (1) | 1 | 0,7 | 0,1–3,6 |
S. Stanley (1) | 1 | 0,7 | 0,1–3,6 |
S. Braenderup (1) | 1 | 0,7 | 0,1–3,6 |
Всего | Total | 153 | 100 | |
Sporadic cases of Salmonella strains isolation | |||
Sporadic incidence (n = 270) | |||
S. Enteritidis | 55 | 20,4 | 16,0–25,6 |
S. Muenchen | 34 | 12,6 | 9,2–17,1 |
S. Typhimurium | 17 | 6,3 | 4,0–9,9 |
Другие серотипы | Other serotypes | 164 | 60,7 | 54,8–66,4 |
Food (n = 239) | |||
S. Infantis | 95 | 39,7 | 33,8–46,1 |
S. Enteritidis | 39 | 16,3 | 12,2–21,5 |
S. Typhimurium | 11 | 4,6 | 2,6–8,1 |
Другие серотипы | Other serotypes | 94 | 39,3 | 33,4–45,6 |
Environment (n = 90) | |||
S. Infantis | 13 | 14,4 | 8,6–23,2 |
S. Typhimurium | 11 | 12,2 | 7,0–20,6 |
S. Bovismorbificans | 10 | 11,1 | 6,1–19,3 |
Другие серотипы | Other serotypes | 56 | 62,2 | 51,9–71,5 |
Total | 599 | ||
543 (72.2%) strains were resistant to at least one of the tested antibiotics, and 193 (25.7%) strains were characterized by multidrug resistance (MDR). The majority of strains with the MDR phenotype (n = 120) were resistant to 4–6 AMA classes, and 6 strains were characterized by the maximum spectrum of resistance — 10 AMA classes out of 11 tested (see Supplementary file in online version on the journal’s website, DOI: https://doi.org/10.36233/0372-9311-451-1).
The percentage of resistant strains of the entire collection of studied Salmonella to certain AMA classes was characterized by the following distribution: 61.3% (95% CI 57.8–64.7) of the studied strains showed phenotypic resistance to quinolones, 28.1% (95% CI 25.0–31.4) — to tetracyclines, 19.1% (95% CI 16.5–22.1) — to penicillins, 18.6% (95% CI 16.0–21. 6) — to β-lactam combination agents, 16.5% (95% CI 14.0-19.3) — to folate pathway antagonists, 10.1% (95% CI 8.2–12.5) — to phenicols, 5.6% (95% CI 4.2–7.5) — to aminoglycosides, 4.7% (95% CI 3.5–6.6)— to cephems, 4.4% (95% CI 3.1– 6.1) — to monobactams, 3.9% (95% CI 2.8–5.6) — to lipopeptides. No penem-resistant strains have been identified.
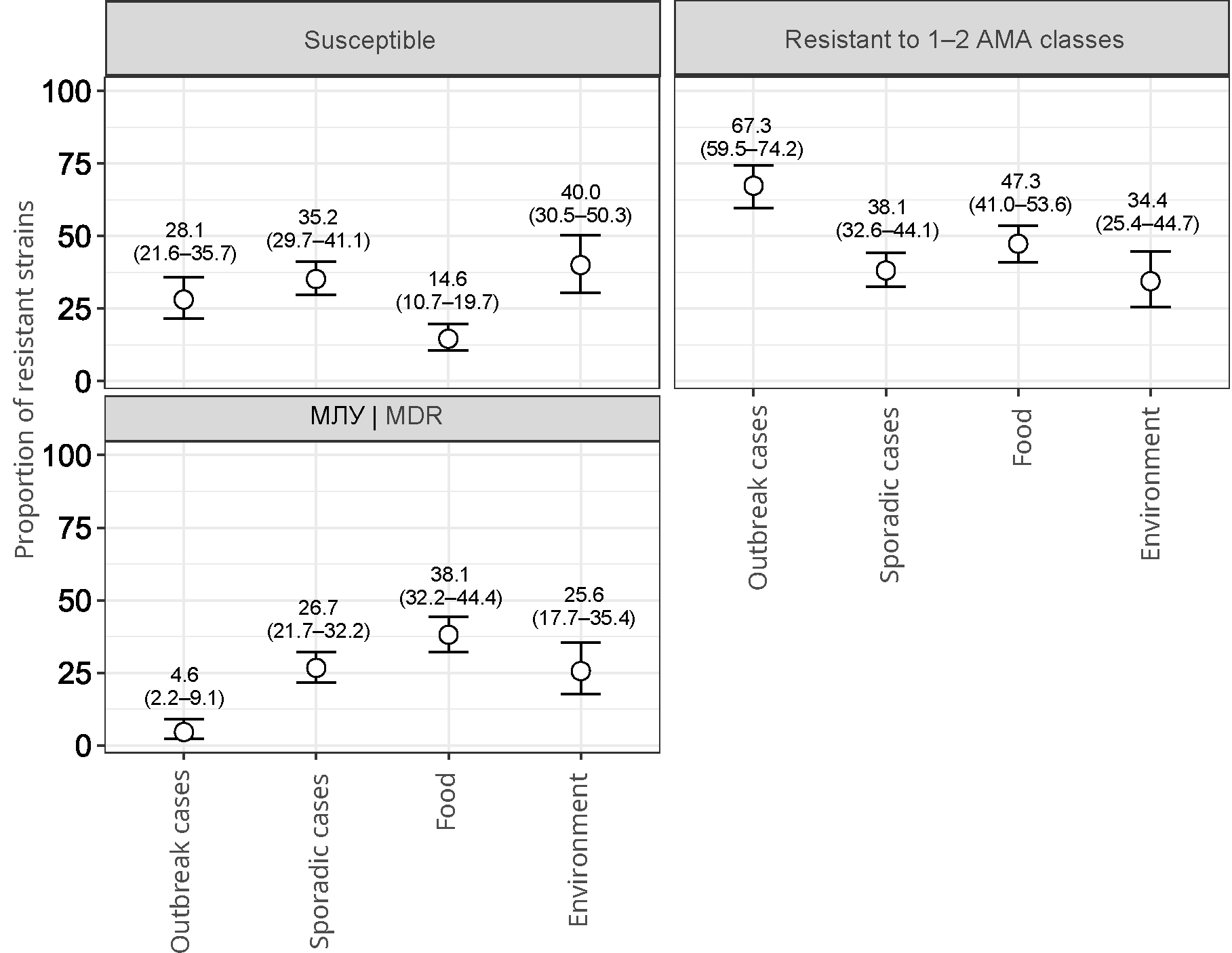
Strains (n = 153) from 113 loci of outbreak incidence of salmonellosis were distributed as follows: in 2019 — 53; in 2020 — 15; in 2021 — 17; in 2022 — 28. 77 (68.1%) outbreaks were characterized by only 1 representative strain. In other cases, 2–4 representative strains were found in the outbreak. The leading serotype in outbreaks was S. Enteritidis (n = 138). In turn, serotypes S. Typhimurium, S. Infantis, S. Muenchen, S. Abony, S. Stanley, S. Braenderup, and S. Bareilly were related to single cases. The largest proportion (67.3%; 95% CI 59.5–74.2) of strains showed resistance to 1–2 AMA classes (Fig. 1). Strains susceptible to all tested AMAs accounted for 28.1% (95% CI 21.6–35.7), while the proportion of MDR strains was only 4.6% (95% CI 2.2–9.1)
Fig. 1. The proportion of resistant Salmonella strains (indicating 95% CI), isolated from various sources and susceptible to all AMAs, resistant to 1–2 AMA classes, resistant to 3 or more AMA classes (MDR).
Isolates obtained from humans as well as from other sources, not associated with cases of outbreak incidence of salmonellosis according to epidemiological data, were represented by 599 strains. Each isolation source had its own dominant serotypes. Among sporadic cases, S. Enteritidis was predominant (20.4%; 95% CI 16.0–25.6), for food and environment — S. Infantis (39.7%; 95% CI 33.8–46.1 and 14.4%; 95% CI 8.6–23.2). At the same time, S. Typhimurium was also among the frequently occurring serotypes (Table 1).
It should be noted that among the strains with sporadic incidence and those isolated from the environment, there were approximately equal proportions of those susceptible to AMAs (35.2%; 95% CI 29.7–41.1 and 40%; 95% CI 30.5–50.3), resistant to 1–2 AMA classes (38.1%; 95% CI 32.6–44.1 and 34.4%; 95% CI 25.4–44.7) and strains with an MDR profile (26.7%; 95% CI 21.7–32.2 and 25.6%; 95% CI 17.7–35.4). Salmonella isolated from food had the highest proportion of MDR phenotypes (38.1%; 95% CI 32.6–44.1) and the lowest proportion (14.6%; 95% CI 10.7–19. 7) of susceptible strains in comparison with other sources (Fig. 1). In addition, strains not associated with outbreak incidence were characterized by a high diversity of antibiotic resistance profiles. We identified 85 different profiles. Frequently occurring serotypes had the largest number of profiles: S. Enteritidis (n = 11), S. Infantis (n = 31), S. Muenchen (n = 10), S. Typhimurium (n = 14), and S. Bovismorbificans (n = 11).
Comparison of Shannon diversity indices (H) based on antibiotic resistance profiles of frequently occurring Salmonella serotypes not related to outbreak incidence, and evaluation of the significance of differences showed that the diversity in S. Infantis strains was significantly higher than in S. Typhimurium strains (p < 0.05; Fig. 2). At the same time, S. Typhimurium did not differ from S. Bovismorbificans in the diversity of resistance profiles (p > 0.05), but had a significantly (p < 0.05) higher diversity than S. Muenchen. The diversity for S. Bovismorbificans showed a slight difference from S. Muenchen (p > 0.05), but was significantly (p < 0.05) higher compared to the diversity of S. Enteritidis. However, S. Muenchen and S. Enteritidis did not differ by diversity (p > 0.05). Thus, S. Infantis had the highest diversity of antibiotic resistance profiles (H = 2.89; 95% CI 2.70–3.08) compared with other serotypes. However, S. Enteritidis had the lowest diversity (H = 1.24; 95% CI 1.01–1.46), despite ranking 2nd by the number of strains isolated from sporadic cases, along with S. Muenchen.
Fig. 2. Distribution of Shannon diversity indices (indicating 95% CI) for Salmonella serotypes not associated with outbreaks and with the largest number of resistance profiles. * — slightly different in diversity (p > 0.05); *** — significant difference (p < 0.05).
Comparative evaluation of phenotypic resistance of Salmonella strains to AMA classes, isolated from various sources
Strains from all sources were characterized by low resistance to aminoglycosides, lipopeptides, and phenicols, while a significant proportion of strains were characterized by high rates of resistance to quinolones (Fig. 3).
Fig. 3. Proportion of resistant Salmonella strains (indicating 95% CI) to different classes of antimicrobial agents from different isolation sources.
Analyzing the frequency of identified resistant strains among various isolation sources relative to certain AMA classes, it can be seen that the proportion of strains resistant to penicillins, β-lactam combination agents, cephems, monobactams, folate pathway antagonists, and tetracyclines was significantly higher or, in case of cephems and monobactams, was found only in the group of sources: “sporadic cases”, “food”, “environment” in comparison with strains from outbreak cases (Fig. 3).
The highest rate of resistance to tetracyclines (49.0%; 95% CI 42.7–55.3) was associated with strains isolated from food products.
The proportion of strains resistant to quinolones and associated with outbreak incidence (68.0%; 95% CI 60.2–74.8) and food products (73.6%; 95% CI 67.7–78.8) was higher compared to other sources.
Comparative evaluation of phenotypic resistance to AMA classes and serotype diversity of Salmonella
Analysis of resistance to AMA classes of strains belonging to the most common Salmonella serotypes revealed the prevalence of resistant strains among the S. Typhimurium and S. Bovismorbificans serotypes. In particular, the proportion of strains resistant to penicillins was 57.1 and 93.9%, and to β-lactam combination agents — 57.1 and 87.9%, respectively. At the same time, the proportion of strains resistant to folate pathway antagonists and tetracyclines in S. Bovismorbificans was at a high level (90.9%; 95% CI 76.4–96.9), and significantly exceeded similar indicators compared to S. Typhimurium (9.5%; 95% CI 3.8–22.1 and 47.6%; 95% CI 33.4–62.3, respectively; Fig. 4).
Fig. 4. Proportion of resistant strains of the most common Salmonella serotypes (indicating 95% CI) to AMA classes.
- Infantis strains differed from other serotypes by their high resistance to quinolones (92.4%; 95% CI 86.2–96.0) and phenicols (24.4%; 95% CI 17.5–32.8), and also showed increased resistance to folate pathway antagonists (45.4%; 95% CI 36.7–54.3) and tetracyclines (76.5%; 95% CI 68.1–83.2).
It should be noted that a significant proportion of strains of all serotypes showed resistance to quinolones and were divided into 3 groups according to the degree of resistance to this AMA class. The first group with the highest proportion of resistant strains included S. Infantis. The second group included S. Enteritidis (70.0%; 95% CI 63.9–75.4) and S. Muenchen (78.0%; 95% CI 64.8–87.2), among which the proportion of resistant strains was significantly higher than for the third group, which included S. Typhimurium (38.1%; 95% CI 25.0–53.2), S. Bovismorbificans (33.3%; 95% CI 19.8– 50.4), and other serotypes (43.7%; 95% CI 37.8–49.6).
The resistance of strains of all analyzed serotypes to cephems, monobactams, and lipopeptides did not exceed 10%. The proportion of strains resistant to aminoglycosides also remained low, less than 10% , and only for S. Infantis (16.8%; 95% CI 11.2–24.5) and S. Bovismorbificans (12.1%; 95% CI 4.8–27.3) this indicator was higher.
Comparison of the diversity of resistance profiles of S. Enteritidis strains isolated from outbreaks and from clinical specimens of sporadic cases
The number of resistance profiles for S. Enteritidis strains isolated from outbreaks (n = 7) was slightly lower than for sporadic incidence (n = 9; Fig. 5). Moreover, all S. Enteritidis strains were characterized by high resistance to quinolones (70.0%; 95% CI 63.9–75.4) and resistance to other AMA classes at a level below 5% (Fig. 4). Only 27.5% (95% CI 22.2–33.5) were sensitive to all antibiotics tested. Comparison of Shannon diversity indices between groups of S. Enteritidis strains isolated from outbreaks (H = 0.98; 95% CI 0.81–1.15) and in sporadic incidence (H = 1.33; 95% CI 1.03–1.62), showed a significant difference (p < 0.05) and a predominance of resistance profiles in the diversity in the case of sporadic incidence (Table 2).
Table 2. The number of Salmonella strains of a certain AMA resistance profile for the compared groups
Resistance profile | Number of strains |
Outbreak cases | |
Susceptible | 35 |
Ciprofloxacin | 90 |
Ampicillin, ampicillin/sulbactam, ciprofloxacin, chloramphenicol | 1 |
Ciprofloxacin, chloramphenicol, tetracycline | 5 |
Chloramphenicol, tetracycline | 2 |
Colistin, ciprofloxacin | 4 |
Tobramycin, ciprofloxacin | 1 |
Total | 138 |
Sporadic cases | |
Susceptible | 20 |
Ciprofloxacin | 26 |
Ampicillin, ampicillin/sulbactam, piperacillin, cefotaxime, ceftazidime, cefepime, aztreonam, ciprofloxacin | 2 |
Ampicillin, ampicillin/sulbactam, piperacillin, cefotaxime, ceftazidime, cefepime, aztreonam | 1 |
Ampicillin, ampicillin/sulbactam, piperacillin, trimethoprim/sulfamethoxazole, ciprofloxacin, tetracycline | 1 |
Ampicillin, ampicillin/sulbactam, piperacillin, trimethoprim/sulfamethoxazole | 1 |
Ciprofloxacin, chloramphenicol | 2 |
Chloramphenicol, tetracycline | 1 |
Tobramycin, ciprofloxacin | 1 |
Total | 55 |
Discussion
Monitoring of phenotypic antibiotic bacterial resistance is an important tool for epidemiological surveillance in the fight against the increase in bacterial resistance to AMAs throughout the world, which is aimed at obtaining information about the dynamics of changes in the phenotypic characteristics of socially significant pathogens and developing comprehensive measures to address this issue. Based on an extensive collection of strains, we attempted to characterize in the presented studies the resistance of non-typhoidal Salmonella circulating in Russia in recent years.
It is necessary to pay attention to the fact that, in order to create a collection of strains for evaluating antibiotic bacterial resistance, we formed a collection of representative Salmonella strains, which, in contrast to a study based on all strains allows us to avoid misconceptions about the detection frequency of a particular resistance profile and obtain objective data. Similar approaches were used before in other studies [17, 18].
An assessment of the serotype diversity of Salmonella and phenotypic resistance to AMA classes showed that the dominant position in terms of isolation frequency was occupied by serotypes S. Enteritidis, S. Infantis, S. Muenchen, S. Typhimurium, S. Bovismorbificans, which accounted for 64.4% of the studied strains. S. Enteritidis strains predominated in the sporadic incidence, and S. Infantis — in food products and environment. In the meantime, S. Typhimurium ranked 3rd by detection frequency in various sources.
The comparison of the levels of resistance to the main AMA classes of the entire collection of strains in our study with the results of previous studies showed a similar picture. According to S.A. Egorova et al., in St. Petersburg and the Leningrad region in 2014–2018 the percentage of resistant strains of non-typhoidal Salmonella averaged 78.6%, and that of strains resistant to quinolones was 63.3% [10], which correlates with the results of our study (72.2 and 61.3%, respectively). At the same time, the proportion of strains resistant to cephems was almost 2 times lower: 1.6% versus 4.7%. Interestingly, the proportion of identified strains with an MDR phenotype was also 2 times lower — 13.0% versus 25.7% in our study, which may be due to differences in the sample of strains relative to the isolation sources. The detection frequency of Salmonella strains with the MDR phenotype in the European Union (28.6%) is close to our average data for the entire collection of strains [19]. According to the report of the National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS),8 the proportion of clinical strains with the MDR phenotype among non-typhoidal Salmonella in the United States was 9.3% in 2018, which is 2 times higher compared to the proportion of strains with MDR, which belonged to outbreak incidence (4.6%) in our study, but is almost 3 times lower when compared with strains found in sporadic incidence (26.7%). The proportion of clinically resistant strains to certain AMA classes, such as quinolones and cephems, identified in the USA was 8.5 and 3.5%, respectively; in our study, these figures corresponded to 68.0 and 0.0% for strains with outbreak incidence, and 51.1 and 7.0% for sporadic cases of disease. The significant discrepancy in quinolone resistance is explained, among other things, by differences in the interpretation criteria of CLSI and EUCAST.
The comparative analysis of Salmonella strains from different sources made it possible to identify a number of patterns. Resistance levels to many AMA classes (penicillins, β-lactam combination agents, folate pathway antagonists, and tetracyclines) were higher in strains not associated with outbreak incidence. This observation is consistent with earlier studies showing that the acquisition of resistance, particularly through horizontal genes transfer, can reduce the adaptiveness and competitiveness of the microorgamisms (fitness of bacteria) having a detrimental effect on the vital physiological processes of the bacterium [20, 21]. Based on our data, this may manifest in a decrease in the virulent properties of Salmonella and, as a consequence, a decrease in the epidemic potential.
The highest percentage of tetracycline-resistant strains was found in food products, and environmental strains had the highest frequency of lipopeptide resistance. The only AMA class to which resistance manifested at a high level across different sources were quinolones. The high frequency of quinolone-resistant Salmonella strains among various sources can be explained by the widespread use of this group of AMAs in animal husbandry. The chemical stability of this group of antibiotics and their ability to accumulate in various environmental objects in low concentrations leads to the emergence and selection of antibiotic-resistant forms of bacteria most effectively [22, 23]. Moreover, chromosomal mutations associated with intermediate-level quinolone resistance in S. Enteritidis do not affect the adaptiveness level of the microorganism [24].
In our study, the percentages of susceptible and resistant strains from sporadic incidence and environmental sources (samples from water bodies and wastewater) were similar, but these isolation sources differed significantly in the diversity of dominant serotypes. According to data published earlier [25, 26], despite the leading position of S. Enteritidis among clinical isolates, this serotype was rarely found in wastewater specimens, while S. Infantis was the dominant serotype in terms of isolation frequency. The data obtained may indicate poor survival rate of S. Enteritidis strains in the environment compared to other serotypes. If we consider the ratio of strains resistant to AMA classes for these isolation sources, we can see that resistance to quinolones, penicillins, β-lactam combination agents and tetracyclines remained at approximately the same level. However, strains isolated from the environment were more resistant to lipopeptides and folate pathway antagonists.
It is noteworthy that the highest percentage of resistant Salmonella strains, including MDR, are associated with food. A significant contribution to the resistance to AMAs for this source was made by S. Infantis strains with their dominant resistance to quinolones (92.4%), tetracyclines (76.5%), folate pathway antagonists (45.4%), phenicols (24.4 %), penicillins (23.5%) and β-lactam combination agents (23.5%). In addition, this serotype had the highest percentage of strains resistant to third-generation cephalosporins (9.2%), which may indicate the production of β-lactamases. The spread of S. Infantis clones with the MDR phenotype is recorded throughout the world [27–29]. According to NARMS, between 2018 and 2019, the number of MDR Salmonella isolates obtained from chicken meat product samples increased from 22% to 29%, while the percentage of MDR isolates from retail chicken meat increased from 20% to 32%9. A significant contribution to the increase in the spectrum of resistance was made by an increase in the number of detected isolates of S. Infantis with the MDR phenotype, which may be a consequence of the use of AMAs such as ampicillin, co-trimoxazole, chloramphenicol, and tetracycline in human and veterinary medicine [11, 30, 31]. The resistance of Salmonella strains isolated from food products to penicillins and tetracyclines in our study (26.8 and 49.0%, respectively) correlates with the data from the European Food Safety Authority (EFSA) [32], in which the resistance of Salmonella strains isolated from broiler and turkey carcasses to ampicillin and tetracycline reached 18.8–19.1 and 48.1–50.3%, respectively.
The analysis of the data obtained allowed identifying the resistance features of clinical strains of Salmonella, which were detected in different epidemiological situations: outbreak and sporadic incidence in people. Thus, outbreaks of salmonellesis were characterized by the highest percentage of strains resistant to 1–2 AMA classes (67.3%) and the lowest percentage of MDR strains (4.6%). On the contrary, the proportion of MDR strains isolated in sporadic incidence was 5.8 times higher than for outbreak incidence, which is explained by the significant contribution of S. Typhimurium strains and other serotypes to the total resistance. Like outbreak strains, sporadic strains were more often resistant to quinolones, which is confirmed by the dominant resistance of the S. Enteritidis (70.0%) and S. Muenchen (78.0%) serotypes to this AMA class. Resistance to penicillins (22.6%), β-lactam combination agents (21.9%), and tetracyclines (21.9%) was significantly influenced by the presence of S. Typhimurium strains, which were more often resistant to these AMA classes. Similar data were published in a joint report by EFSA and the European Center for Disease Prevention and Control in 2022 [32], which stated that a high percentage of MDR Salmonella strains (25.4%) isolated from humans in 2019–2020 was justified by the significant contribution of S. Typhimurium and S. Kentucky resistant strains. High levels of resistance to ampicillin (29.8%), sulfonamides (30.1%), and tetracyclines (31.2%) varied by serotype from low for S. Enteritidis to extremely high for S. Typhimurium and S. Kentucky.
Over the period of many years of monitoring, S. Enteritidis remains the main serotype causing outbreak incidence of salmonellosis [33]. Our data demonstrate that both among outbreak and sporadic cases of salmonellosis, the dominant serotype was S. Enteritidis. In addition, this serotype occupied the second ranking position (16.3%) when isolated from food products. Comparison of S. Enteritidis strains from outbreak and sporadic cases showed a slight difference in the number of resistance profiles and a low level of resistance to many AMA classes, except for quinolones. However, despite the higher number of strains from outbreaks (2.5 times higher than sporadic ones), the diversity of resistance profiles was significantly lower. Another feature of S. Enteritidis is the high rate of occurrence of strains with resistance to quinolones, which amounted to 70.0%, which is consistent with the data from studies conducted in St. Petersburg [10], where the resistance of S. Enteritidis strains isolated from humans was 71.0%. According to the NARMS surveillance system10, S. Enteritidis strains were the most common serotype among isolates with reduced ciprofloxacin susceptibility in 2019, accounting for 45% of Salmonella isolated from humans. It is known that resistance to quinolones in Salmonella can be caused by the presence of horizontally (plasmids, transposons, integrons) and vertically (chromosomal mutations) inherited genetic traits [34]. It was shown that the contribution of chromosomal mutations to quinolone resistance for S. Enteritidis was significantly higher [35]. The percentage of Salmonella strains resistant to quinolones due to single nucleotide substitutions in the region of the DNA gyrase and topoisomerase IV genes reached 97.54%, while the presence of plasmid-mediated resistance was detected only for 1.1% of strains [36].
The second rank by the number of strains isolated from humans was occupied by the S. Muenchen serotype (12.6%). It was also the etiological agent in two outbreaks of salmonellesis during the four-year observation period. An increase in the incidence of this serotype is recorded both in individual regions of Russia [37, 38] and in other countries [39]. According to the Salmonella Monitoring Reference Center11 for 2021, S. Muenchen occupied the 5th ranking position in terms of frequency of isolation from humans and food products and the 6th position — from the environment. It is interesting
to note that in our study, S. Enteritidis and S. Muenchen had the least diversity of resistance profiles and did not differ significantly from each other.
Conclusion
In the presented study, we have described the phenotypic resistance of Salmonella and compared the diversity of resistance profiles of the most common serotypes in Russia, based on a sample of representative strains of non-typhoidal Salmonella. A significant contribution to the population of resistant Salmonella is made by strains that are not related to the outbreak incidence. Almost half (57.6%) of the studied strains were resistant to at least one AMA class, among which the most frequently detected resistance was to quinolones, tetracyclines, penicillins, and folate pathway antagonists, which have been applied for a long time or are currently used in human and veterinary medicine.In addition, non-typhoidal Salmonella isolated from various sources had a wide variety of resistance profiles. Among them, the largest number of strains characterized by resistance were found among serotypes S. Infantis, S. Typhimurium, and S. Bovismorbificans.
Continuous monitoring of antibiotic resistance and tracking of MDR in Salmonella strains plays a key role in addressing the issue of antibiotic resistance and provides valuable information for epidemiological surveillance in order to develop prevention strategies.
Supplemental information to the article
1 On the state of sanitary and epidemiological well-being of the population in the Russian Federation in 2022: State report. Moscow; 2023.
2 Plumb I., Fields P., Bruce B.B. Salmonellosis (Non-typhoidal). CDC Yellow Book; 2024. Travel-Related Infectious Diseases.
URL: https://wwwnc.cdc.gov/travel/yellowbook/2020/travel-related-infectious-diseases/salmonellosis-non0typhoidal (access date 7/9/2023).
3 Clinical guidelines. Salmonellosis in adults. M.; 2021.
4 The WHO Essential Medicines List Antibiotic Book: improving antibiotic AWaReness (Draft for consultation). Geneva; 2022.
5 Global antimicrobial resistance and use surveillance system (GLASS) report 2022. Geneva; 2022.
6 Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI supplement M100 Clinical and Laboratory Standards Institute, USA, 2021.
7 Integrated Surveillance of Antimicrobial Resistance in Foodborne Bacteria: Application of a One Health Approach. Geneva; 2017.
8 Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2019. Atlanta; 2023.
9 Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2019. Atlanta; 2023.
URL: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/2019-narms-update-integrated-report-summary (дата обращения 07.09.2023)
10 Centers for Disease Control and Prevention. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): human isolates final report, 2019. Atlanta; 2023.
11 Rozhnova S.Sh., Podkolzin A.T., Kuleshov K.V. et al. Information Bulletin of Salmonella Monitoring Reference Center No. 34. Moscow; 2022.
About the authors
Anastasia S. Pavlova
Central Research Institute of Epidemiology
Author for correspondence.
Email: a.pavlova@cmd.su
ORCID iD: 0000-0003-4619-9337
researcher, Laboratory of molecular diagnostics and epidemiology of enteric infections, Central Research Institute of Epidemiology
Russian Federation, MoscowKonstantin V. Kuleshov
Central Research Institute of Epidemiology
Email: a.pavlova@cmd.su
ORCID iD: 0000-0002-5238-7900
Cand. Sci. (Biol.), Head, Laboratory of molecular diagnostics and epidemiology of enteric infections, Central Research Institute of Epidemiology
Russian Federation, MoscowNatalia E. Krutova
Central Research Institute of Epidemiology
Email: a.pavlova@cmd.su
ORCID iD: 0000-0003-2925-5376
junior researcher, Laboratory of molecular diagnostics and epidemiology of enteric infections, Central Research Institute of Epidemiology
Russian Federation, MoscowAnna N. Guseva
Central Research Institute of Epidemiology
Email: a.pavlova@cmd.su
ORCID iD: 0000-0001-7028-0253
junior researcher, Laboratory of molecular diagnostics and epidemiology of enteric infections, Central Research Institute of Epidemiology
Russian Federation, MoscowAlexandr T. Podkolzin
Central Research Institute of Epidemiology
Email: a.pavlova@cmd.su
ORCID iD: 0000-0002-0044-3341
D. Sci. (Med.), Deputy director for epidemiology, Central Research Institute of Epidemiology
Russian Federation, MoscowReferences
Supplementary files