Live attenuated COVID-19 vaccines: approaches to development and prospects for clinical use
- Authors: Korchevaya E.R.1, Gracheva A.V.1, Dyakov I.N.1, Zverev V.V.1, Faizuloev E.B.1,2
-
Affiliations:
- I. Mechnikov Research Institute for Vaccines and Sera
- Russian Medical Academy of Continuous Professional Education
- Issue: Vol 100, No 3 (2023)
- Pages: 225-236
- Section: REVIEWS
- URL: https://microbiol.crie.ru/jour/article/view/15838
- DOI: https://doi.org/10.36233/0372-9311-404
- EDN: https://elibrary.ru/psdxzr
- ID: 15838
Cite item
Abstract
Although WHO declared an end to the pandemic, COVID-19 remains a significant public health concern worldwide. Modern vaccines often induce either only humoral or only cellular immunity. Furthermore, new emergent epidemiologically significant SARS-CoV-2 variants and their spread considerably reduce the effectiveness of preventive vaccination. Therefore, there is an urgent need to improve the existing vaccines against COVID-19. One of the promising approaches to the solution of the problem is creation of a "universal" vaccine that would have a cross-protective activity against different antigenic variants of the virus. In this respect, the development of live attenuated vaccine is of special interest, as it can activate not only humoral, but also cell-mediated components of immunity, providing long-term immune response and cross-protection against different variants of the virus.
This review highlights the existing approaches to producing attenuated SARS-CoV-2 strains and gives an assessment of their prospects for clinical use. Some researchers use methods of genetic engineering and reverse genetics such as site-directed mutagenesis and codon deoptimization for virus attenuation. Others tend to use traditional approaches focusing on producing virus mutants through extended passaging in cell culture under selective conditions. The gained experience demonstrates great prospects for development of highly effective live-attenuated vaccine against COVID-19.
Keywords
Full Text
Introduction
Although WHO declared an end to the pandemic, COVID-19 remains a significant public health concern worldwide. Efforts unprecedented by their scale are being taken to reduce the burden of this disease, including mass vaccination, development of new and "retargeting" of existing pharmaceutical products. Viral vector, mRNA, recombinant protein subunit vaccines and inactivated whole-virion vaccines have been developed worldwide and are used for specific COVID-19 prevention [1–5]. The application of these technology platforms shortens the time required for development of vaccines with high safety profile, helps prevent severe COVID-19 cases or fatal outcomes caused by a homologous variant of the virus (i.e. the virus variant, based on which the vaccine is developed). The emergence and wide spread of new epidemiologically significant SARS-CoV-2 variants, especially the Omicron variant and its sublineages evading the immune protection developed against the Wuhan strain that was used in development of most of the approved vaccines, have considerably reduced the effectiveness of preventive vaccination [6–8]. Therefore, it is advisable to improve the existing vaccines and their match with the circulating SARS-CoV-2 variant to maintain the effectiveness of vaccination at high levels. One of the promising approaches to the solution of the problem is creation of a "universal" vaccine that would have a cross-protective activity against different antigenic variants of the virus. For example, good prospects are offered by the development of a live-attenuated vaccine (LAV) capable of activating not only humoral, but also cell-mediated components of immunity, providing long-term immune response and cross-protection against different variants of the virus [9, 10]. It should be noted that currently the potential of LAVs in prevention of COVID-19 remains almost unrealized.
Currently approved LAVs are highly effective and are successfully used for specific prevention of such diseases as poliomyelitis, measles, rubella, chicken pox, mumps, and influenza [11]. These vaccines were developed by obtaining mutant forms of the virus, which have reduced ability to replicate in human cell culture [12, 13], or by creating cold-adapted (ca) temperature sensitive (ts) variants incapable of replication at human body temperature [14].
The purpose of this review is to highlight the existing approaches to SARS-CoV-2 attenuation and to assess the prospects for clinical use of LAVs against COVID-19.
Approaches to SARS-CoV-2 attenuation
Certain experience has been accumulated in producing attenuated SARS-CoV-2 strains. Some researchers use methods of genetic engineering and reverse genetics such as site-directed mutagenesis and codon deoptimization for virus attenuation [15–20]. Others tend to use traditional approaches focusing on producing virus mutants through extended passaging in cell culture under selective conditions [21–24]. Currently, the only LAV against COVID-19 - CoviLiv™ vaccine (previously called COVI-VAC; developers: Codagenix (United States) and the Serum Institute (India) is undergoing clinical trials; it is a virus with deoptimized codons in the S protein [5].
Classical (traditional) approaches to SARS-CoV-2 attenuation
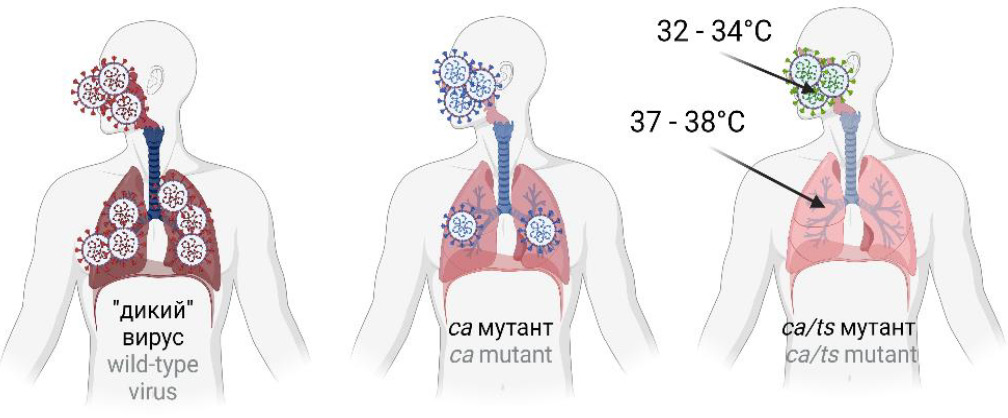
One of the classical approaches to virus attenuation is virus adaptation to growth at low temperatures; usually, it is performed in cells of a different host. It was used for development of live vaccines against influenza, rubella, and measles [25–29], and such vaccines are widely used in different countries of the world. Cold adaptation is a process, by which the virus gradually adapts to growth in cell culture at low temperatures; it is often associated with reduced replication at 37ºC and higher temperatures. The adaptation of the virus to a new host and to the growth at low temperatures results in development of mutant forms of the virus, which are characterized by decreased replicative activity in a human body. For example, the virus having ca- and ts-phenotypes is not able to replicate effectively and to cause pathological changes in the human body where the temperature is higher than 37ºC (Fig. 1). The ca-phenotype represents the ability of the virus to replicate effectively in cell culture at low temperatures compared to the parent strain. The ts-phenotype is characterized by the inability of the virus to replicate at 37ºC, 39ºC or 41ºC, while these temperatures are favorable for replication of the parent strain.
Fig. 1. Virulence of the wild-type, cold-adapted (ca) and temperature-sensitive (ca/ts) strains of the respiratory virus. The wild-type strain can affect the upper and lower respiratory tract, causing pneumonia, while the ca-strain primarily affects the upper compartments, and ca/ts affects only the upper compartments of the respiratory tract.
For illustrative purposes, we can refer to the existing analog vaccine against the respiratory virus, namely the live-attenuated influenza vaccine (LAIV), which is administered intranasally. LAIV induces development of not only systemic humoral and cell-mediated immunity, but also local mucosal immunity at the point of entry of the virus into the body (the "entrance gate" of infection) – the respiratory mucosa. LAIV is based on using the ca-strain – an attenuation donor bearing in "internal genes" mutations, which are responsible for the ts-phenotype of the virus (the ability to replicate at 25–33ºC, but not at 37–39ºC) [30]. "Internal genes" encode all proteins of the influenza virus except for surface antigens –hemagglutinin and neuraminidase. Thus, when administered intranasally, ca/ts-strains of the influenza virus affect the upper respiratory tract (this specifies their immunogenicity), but they are not able to replicate in lungs and cause pneumonia. Vaccine strains for LAIV are obtained by reassortment between the donor strain and circulating strains, following which reassortant strains carry "internal genes" responsible for their attenuation (att) phenotype as well as hemagglutinin and neuraminidase genes responsible for protective properties of the virus [29–31]. Drawing on the experience gained in the LAIV development, several groups of researchers are working on LAV against COVID-19.
For SARS-CoV-2 attenuation, in some studies, the virus was adapted to growth at low temperatures combined with change of the host [21, 22, 24, 32], while Li et al. achieved attenuation solely by changing the host [13]. The isolation and further cultivation of the virus were performed in monkey kidney cell culture (the Vero CCL-81 or Vero E6 cell line). To obtain ca-strains, the wild-type (parent) strain was passaged at the temperature gradually decreased to 21–23ºC. When the desired temperature was reached, the ca-virus went through a few more passages at the low temperature to secure the phenotype and was cloned using the plaque method [24] or the limiting dilution method [21, 32]. The randomly selected ca-clones obtained during the previous stage were examined for the presence of the ts-phenotype [21, 24, 33], which, like the ca-phenotype, is a possible marker of the virus attenuation [14].
Seo et al. were the first to describe the development of the attenuated cold-adapted SARS-CoV-2 virus [21]. The clinical isolate of SARS-CoV-2 was attenuated by cold adaptation to 22ºC; each passage was performed at a lower temperature if pronounced CPE was demonstrated. The obtained attenuated strain had both ca- and ts-phenotypes, which was associated with 59 mutations present in genes of structural and nonstructural proteins, including 37 nonsynonymous substitutions. The K18-hACE2 transgenic mice expressing the gene of the human ACE2 receptor were immunized with the attenuated virus; after they were experimentally infected with the wild-type SARS-CoV-2, they not only did not die, but also did not have any clinical signs of the disease compared to the animals from the control group. The att-phenotype of the ca-virus was confirmed by the absence of pronounced histopathology in mouse lungs and reduced replication in the internal organs on the 6th day after the immunization compared to the wild-type virus.
The study performed at the Mechnikov Research Institute of Vaccines and Sera was focused on the attenuation of the Dubrovka (D) strain of SARS-CoV-2 by cold adaptation (gradual decrease of the growth temperature during 42 passages to 23ºC) in Vero CCL-81 cells [32]. Two ca-clones of the D strain were obtained: The D-D2 variant having the ts-phenotype and capable of replication in Calu-3 human lung cells; and the D-B4 variant that did not have the ts-phenotype and lost the ability to replicate in Calu-3 human lung cells. The genome-wide genetic characterization of the D-D2 and D-B4 variants revealed up to 20 nucleotide and 18 amino acid substitutions compared to the parent strain. In the intranasally infected Syrian hamsters, both ca-variants demonstrated reduced virulence; they did not slow down the weight gain, significantly more slowly replicated in the lungs and other organs, caused significantly less pronounced inflammatory changes in the lungs compared to the D strain. Thus, it was concluded that for ca-variants of SARS-CoV-2, the ts-phenotype was not critical for virulence reduction. For example, the D-B4 variant that did not have the ts-phenotype and lost its ability to infect Calu-3 human lung cells also demonstrated the reduced virulence in hamsters [23].
Xu et al. attenuated SARS-CoV-2 by passaging the virus in the Vero E6 cell culture at the gradually decreased temperature. When the temperature of 21ºC was reached, 5 passages were carried out, followed by cloning and assessment of the temperature sensitivity. Finally, the TS11 clone was chosen as having ca- and ts-phenotypes. The whole-genome analysis of the TS11 clone revealed the deletion leading to the loss of a 12 amino-acid region of the protein in the furin cleavage site in the S protein, and the 371-nucleotide deletion involving ORF7b-ORF8 genes as well as several point mutations in nsp16, S, E, orf7a and N genes. After the intranasal infection with the TS11 clone, the Syrian hamsters continued to gain weight and did not demonstrate any signs of the disease. TS11 successfully replicated in the nasal cavity, but not in the lungs, causing only minor lesions in them. By the 20th day after the immunization, the histological analysis did not detect any traces of inflammation in the lungs [24].
Abdoli et al. also obtained the SARS-CoV-2 ca-mutant by passaging the virus in the Vero cell culture at the gradually decreased temperature. The researchers assume that the attenuation of the obtained KaraVac strain was caused by the double deletion in the S protein: at the S1/S2 junction (the PRRA motif) and at the S1-NTD site (the GTNGTKR motif). The KaraVac strain replicated at 25ºC, 33ºC and 39ºC, though not at 41ºC. Compared to the control group infected with the wild-type strain, the immunized Syrian hamsters did not have any weight loss and did not have any other signs of the disease [22].
At the same time, Li et al. developed an attenuated variant of SARS-CoV-2 by performing serial passages in the Vero cell culture at 37ºC to obtain material for an inactivated vaccine. The developed strain was called VAS5 and had a deletion, which consisted of 21 nucleotides encoding 7 amino acids, and was located upstream of the S1/S2cleavage site in the S gene. In the Caco-2 and Huh7 cell cultures as well as in human lung epithelial organoid cells, the VAS5 strain replicated significantly more slowly than the wild-type strain. The immunized K18-hACE2 mice did not have any weight loss; no pathological changes were detected in the lungs; only few RNA copies were detected in the lung homogenates [13].
Genetic engineering-based approaches to SARS-CoV-2 attenuation
The development of an attenuated virus by using classical virological methods can take several months, thus posing a problem in the pandemic situation. The advances in genetic engineering promoted new approaches to fast development of vaccine strains using site-directed mutagenesis [34–36] or codon deoptimization [37, 38]. For example, the studies of Murine coronavirus (previously known as Murine hepatitis virus), which is a well-studied model coronavirus, have found that sequences of nonstructural NSP14 and NSP16 proteins, which are highly conserved for different coronaviruses, are universal targets for site-directed mutagenesis aimed at reduction of virus virulence. Even a single amino acid substitution resulted in pronounced virus attenuation in mice. At the same time, immunization of the mice with the mutant virus promoted development of a long-term humoral and robust CD4+- and CD8+-T-cell-mediated immune response as well as full protection against infection with a lethal dose of the virus [39].
Ye et al. also used the SARS-CoV-2 NSP16 gene as a target, introducing the D130A mutation into it. The NSP16 protein is a type I interferon antagonist and is critically important for methylation of the 5' terminal cap structure of viral mRNAs, while the D130A point mutation leads to inactivation of the NSP16 methyltransferase activity. The d16 mutant strain during the extended passaging in the Vero E6 cell culture remained genetically stable and did not demonstrate any signs of virulence reversion. The Syrian hamsters and K18-hACE2 mice experimentally infected with the d16 strain did not demonstrate any weight loss and did not develop the disease [20].
Liu et al. used genes of SARS-CoV-2 accessory proteins as a target, as they are associated with the regulation of virus replication through interaction with host signaling pathways. Genes of the ORF3, ORF6, ORF7 and ORF8 accessory proteins were removed from the genome, and the TRS ACGAAC fragment of the transcription regulatory sequence was replaced with CCGGAT. The resulting attenuated ∆3678 virus replicated less efficiently in the primary culture of human lung epithelial cells compared to the parent strain, though it retained its replication ability in the Vero E6 cell culture. In the tests using BALB/c mice, the role of each gene in the SARS-CoV-2 attenuation was studied using the SARS-CoV-2 strain adapted to mice. It was found that the ORF3 (∆3a) deletion played an important role in the attenuation of the ∆3678 virus, which could be explained, as assumed by the researchers, by the impact of the ORF3 product on the signaling pathway of the type I interferon. The Syrian hamsters immunized intranasally with the ∆3678 strain did not lose weight; the viral load in nasopharyngeal, tracheal and lung washes was significantly lower than the viral load in the control group infected with the wild-type strain. After the immunization, The K18-hACE2 mice did not lose weight and did not die even after they had received a high dose of the attenuated virus [18]. The similar results were obtained by Ye et al. in their studies [20].
To obtain an attenuated strain, Liu et al. made modifications in the SARS-CoV-2 (the WA1/2020 isolate) genome: They removed the sequence encoding the PRRA peptide upstream of the furin cleavage site, removed ORFs6-8s, which are interferon antagonists, introduced K164A/H165A mutations into the C-terminal domain of the NSP1 protein. The developed strain WA1-ΔPRRA-ΔORF6-8-Nsp1N128S/K129E demonstrated poorer replication than the original virus in the MatTek EpiAirway human tracheal and bronchial epithelial cell culture and caused only mild lung lesions in infected K18-hACE2 transgenic mice and Syrian hamsters [19].
A slightly different approach was used by Yoshida et al. [17]. Using reverse genetics methods, they obtained a library of 659 mutant clones based on the clinical isolate of SARS-CoV-2 B.1.1 lineage (Pango). To identify the ts-phenotype, they grew clones using two temperature regimes – 32ºC and 37ºC, selecting clones that did not have a cytopathic effect at 37ºC for further tests. The experimental infection of Syrian hamsters also confirmed the att-phenotype of the selected ts-strains, which was characterized by the absence of weight loss, reduced replicative activity of the virus in the respiratory tract, less pronounced pneumonia compared to the original strain [17].
For the SARS-CoV-2 attenuation, some researchers used the codon deoptimization technique involving replacement of original codons with suboptimal ones while preserving amino acid sequences of viral proteins [38]. Such alterations have a direct effect on the rates of viral protein production and genome replication, being responsible for attenuation of the virus [38]. Using this technique, Trimpert et al. recoded most of the SARS-CoV-2 genomic sequence and obtained several candidate vaccine strains. Codons that can be rarely found in the human genome were selected for deoptimization. The sCDP9 and sCDP10 strains selected for further tests replicated more poorly in the cell culture than the original strain and remained genetically stable during 10 passages in the Vero E6 cell culture. The intranasally immunized Syrian hamsters had minor losses in weight, but quickly regained it; they did not demonstrate any signs of the disease, had less significant inflammatory changes in the lungs [15].
Using the codon deoptimization technique, Codagenix (United States) developed the CoviLiv™ vaccine [16]. The S gene served as a target for deoptimization. After the immunization with the vaccine strain, Syrian hamsters did not lose weight; the histological examination of the lungs on the 6th day detected only minor inflammatory changes. The infectious titer in the lungs of hamsters was measured at the limit of detection for this technique, thus being indicative of extremely low replicative activity of the vaccine strain in the lungs.
The Table summarizes the experience of different research groups in the development of attenuated variants of SARS-CoV-2.
Methodological approaches to SARS-CoV-2 attenuation after intranasal administration
Strain name | Attenuation strategy | Animal model on which the att phenotype was established | Reference |
D-D2 и D-B4 | Cold adaptation in Vero cell culture up to 23ºС | Syrian golden hamsters | [23] |
CoV-2-CNUHV03-CA22⁰C | Cold adaptation in Vero cell culture up to 22⁰С | K18-hACE2 mice | [21] |
Δ3678 | ORF3, ORFs6-8 deletion, changing ACGAAC to CCGGAT in TRS | K18-hACE2 mice, Syrian golden hamsters | [18] |
KaraVac | Cold adaptation in Vero cell culture up to 25ºС | Syrian golden hamsters | [22] |
sCPD9 и sCPD10 | Codon-pair deoptimization | Syrian hamsters, Roborovski dwarf hamster | [15] |
VAS5 | Serial passages in Vero cell culture | K18-hACE2 mice, hamsters | [13] |
TS11 | Cold adaptation in Vero cell culture up to 21ºС | Syrian golden hamsters | [24] |
CoviLiv (COVI-VAC) | Codon-pair deoptimization of S-gene | Syrian golden hamsters | [16] |
rTS-all | Random mutagenesis and generation by reverse genetic methods strain with all nessessary ts-related mutations | Syrian golden hamsters | [17] |
WA1-ΔPRRA-ΔORF6-8-Nsp1K164A/H165A | ORFs6-8 deletion, removing PRRA upstream of the furin cleavage site, introducing K164A/H165A into the C-terminus of NSP1 | K18-hACE2 mice, Syrian golden hamsters | [19] |
d16 | Point mutation D130A in NSP16 gene | K18-hACE2 mice, Syrian golden hamsters | [20] |
Immunogenic potential of live-attenuated vaccines against COVID-19
The protective activity demonstrated by most of the licensed vaccines against COVID-19, which are designed to develop humoral immunity, depends on induction of neutralizing antibodies to nonstructural surface S protein of SARS-CoV-2 (Fig. 2, a). However, the pressure of artificial selection induced by mass immunization makes this target highly variable, thus leading to virus evasion of immune surveillance and a rapid decrease in the efficacy of vaccines against re-emerging variants of SARS-CoV-2 [6, 7]. These vaccines do not provide sterilizing immunity [40] and robust mucosal immunity [41].
Fig. 2. Mechanisms of induction of an immune response by the intramuscular immunization vaccine containing the S protein and by intranasal vaccination of LAV. a — vaccines against COVID-19, which are based on the S protein of SARS-CoV-2 (using the example of the viral vector vaccine), induce only a humoral response involving production of virus-neutralizing antibodies; b — LAV triggers the mechanisms inducing acquired immunity, which are similar to those engaged in natural infection, including activation not only humoral, but also cell-mediated immunity. In intranasal administration, the vaccine strain infects epithelial cells of the upper respiratory tract, thus inducing both local (mucosal) and systemic immune responses. The established protection including cellular and humoral components provides immunity both at the level of the entrance gate of the infection (mucosa) and at the systemic level. i/m — intramuscular injection; i/n — intranasal; APC — antigen-presenting cell; CTL — cytotoxic lymphocyte; MHC — major histocompatibility complex.
In natural infection with SARS-CoV-2, the immune response is developed against all viral proteins — both structural and nonstructural. The M and N structural proteins are highly immunogenic and, together with nonstructural proteins, are more conserved than the S protein. In addition, many T-cell epitopes of phylogenetically related species of coronaviruses and different variants of SARS-CoV-2 are not located in the S protein [42, 43]. If an infected organism develops an effective T-cell-mediated immune response, they do not develop a systemic excessive inflammatory response, thus presenting a mild, sometimes asymptomatic case. If the development of the T-cell-mediated (and consequently humoral) response is delayed, the dominant role is played by factors of the innate immunity, being manifested at the systemic level and causing a severe disease [44]. It should be noted that the protectivity of the T-cell-mediated immunity depends less on mutations that occur in new variants of SARS-CoV-2, as it is targeted at more conserved viral antigens [45, 46].
The protective activity of live-attenuated vaccines for intranasal application (mucosal vaccines) employs the same mechanisms that participate in the development of acquired immunity during natural infection (Fig. 2, b). When the respiratory virus enters nasal and/or oral cavities, the lymphoid tissues associated with the mucosa of the respiratory and digestive tract turn into the first line of defense against viral infection. This process involves all components of innate upper respiratory immunity - cellular (neutrophils, macrophages, dendritic cells, resident microfold M cells, innate lymphoid cells, natural killer cells and mast cells) as well as soluble molecules (galectins, collectins, cytokines, etc.). The activation of the first line of defense is followed by activation of dendritic cells in the focus of infection; they take up, process and present viral antigens. Such activated dendritic cells "loaded" with viral antigens migrate to regional lymph nodes; antigens are presented to naïve T cells, thus triggering their further differentiation and activation of adaptive immunity, which is accompanied by production of cytotoxic T cells and helper T cells. Specific naïve B cells recognize viral antigens either without any assistance or when they are presented by follicular dendritic cells as an antigen-antibody complex in a Fc-receptor-connected manner. The interaction of activated helper T cells and B cells triggers the humoral immune response. The simultaneous expansion of CD4+ helper T cells, CD8+ cytotoxic T cells in the focus of infection and production of antibodies by plasma cells is of critical importance for elimination of the virus [41, 47, 48]. The combined activation of humoral and cellular components of the systemic and mucosal immune protection can provide effective protection against infection with SARS-CoV-2 [47].
LAVs are administered in different ways: vaccines against measles, rubella, mums and chicken pox are administered intramuscularly; polio and rotavirus vaccines are given by mouth; LAIV is administered intranasally. The developers of LAVs against COVID-19 use the intranasal administration [15–24]. Different routes of administration have their advantages and disadvantages. For example, intranasal and oral administration provides not only induction of systemic cell-mediated and humoral adaptive immune responses, but also mucosal (local) immunity, including secretion of specific IgA antibodies in the mucosa of the respiratory tract or intestines. In intranasal immunization followed by infection with a virulent strain, specific secretory IgA antibodies neutralize the virus directly in respiratory mucosa, which is the "entrance gate" of the infection, by suppressing its adhesive ability and reducing transmission efficiency [47]. At the same time, if intranasal immunization is performed using an insufficiently attenuated virus, there is a risk of damage that may be caused to the central nervous system through olfactory nerves [48]. For LAVs administered intramuscularly, there is no risk of brain damage; after the second dose of the vaccine, the seroconversion rate reaches 95–100% [49, 50]. Mehla et al. found that when the vaccine strain of SARS-CoV-2 was administered intranasally, the titer of neutralizing antibodies in serum was significantly higher than after intramuscular administration [51].
While, according to WHO1, the CoviLiv™ vaccine is going through phase III of clinical trials, no information is available regarding the safety and efficacy of LAV against COVID-19 for people. At the same time, the official website of the vaccine developer has already announced that the requested report is going to be published soon.2 The website confirms that the live-attenuated intranasal vaccine against COVID-19 CoviLiv™ is immunogenic, well tolerated by healthy adults, and induces protective cell-mediated immunity against all the known variants of SARS-CoV-2. A total of 48 healthy adult individuals took part in the clinical trials. The company press release informs that the clinical trials for the vaccine are included in the WHO-sponsored program – Solidarity Trial Vaccines, which is aimed to support development of second-generation COVID-19 vaccines having a higher efficacy toward new epidemiologically significant variants, longer protection, more simple storage requirements and a needle-free method of immunization.
The immunogenicity and protective activity of LAVs have been extensively studied only in animal models for coronavirus infection. The main models for preclinical studies of attenuated mutants of SARS-CoV-2 were golden Syrian hamsters (Mesocricetus auratus), Roborovski dwarf hamsters (Phodopus roborovskii) and K18-hACE2 transgenic mice. In most of the studies, immunized animals developed a protective immune response against the parent strain SARS-CoV-2, which was used for development of an attenuated strain. Syrian hamsters immunized with an attenuated virus and then experimentally infected with the parent strain did not lose weight and did not demonstrate any clinical signs of the disease; the titration of nasal washes and long homogenates showed lower titers of the virus compared to the animals from the control group [17, 18, 22, 23]. Trimpert et al. noted that on the 2nd, 3rd and 5th day after the experimental infection, no infection activity was detected in the lung samples from the immunized animals, thus confirming the establishment of sterilizing immunity [15, 52]. Xu et al. also reported that the hamsters immunized with the ts-strain did not demonstrate any signs of infection, even being accidentally cross-infected by the animals infected with the wild-type strain [24]. Liu et al. pointed out that the single-dose intranasal immunization of the Syrian hamsters with the Δ3678 att-strain of SARS-CoV-2 not only protected them against infection with the virulent strain, but also decreased the risk of virus transmission [18]. During their studies, Seo et al. found that the K18-hACE2 mice immunized with the ca-strain not only remained alive after the experimental infection, but also did not lose weight and did not have any clinical signs of the disease [21].
Li et al. also assessed the risk of transmission of the SARS-CoV-2 virus from the previously immunized animals after they were infected. On the next day after the infection, the immunized hamsters were put into cages with the hamsters that had not been previously immunized and had not been exposed to the virus; washes were collected on the 3rd and 5th day. On the 3rd day, in non-immune hamsters, viral RNA was detected at the PCR detection limit; on the 5th day, RNA was not detected, thus being indicative of the absent virus transmission through the close contact with the immunized and infected hamsters [13].
At the same time, Liu et al. pointed out possible limitations for immunogenicity assessment using K18-hACE2 mice-based models: Encephalitis caused by the infection with att-SARS-CoV-2, which is followed by the death of animals, makes it difficult to interpret the results. To address the above limitation, nasal, lung and brain samples were collected on the 2nd, 4th and 6th day after the infection to measure the viral load. Curiously, even the most attenuated strain WA1-ΔPRRA-ΔORF6-8-Nsp1N128S/K129E remained neurotropic and was detected in the brain of the K18-hACE2 mice at high levels on the 6th day after the infection [19]. In the studies performed in Syrian hamsters, att-strains of SARS-CoV-2 did not demonstrate such high neurotropism and pathological changes in the brain.
LAVs against SARS-CoV-2 have a high potential of cross-protective activity against different variants of the pathogen, as the same mechanisms that are employed in natural infection with the virus are put into action. The natural infection with SARS-CoV-2 prevented up to 90% of re-infection cases with Alpha, Beta or Delta variants [53, 54] and 56% of re-infection cases with the Omicron variant [55]. Most of the re-infection cases occurred one year after the primary disease [55]. Re-infection has become a common thing since the emergence of the Omicron variant and its further evolution. The study performed in Qatar demonstrates that the infection of people with SARS-CoV-2 variants preceding the omicron variant provided less than 60% of protection against re-infection with Omicron subvariants. At the same time, during re-infection with the Omicron variant, the protective effectiveness of the primary infection against development of severe disease forms or death reached 97.3% (95% CI, 94.9–98.6%) regardless of the virus variant that had caused the primary infection [56]. The extremely low percentage of severe and fatal cases after re-infection offers hope that LAVs against COVID-19 will be able to provide cross-protection against different variants of the virus.
Trimpert et al. demonstrated experimentally that when infected with att-strains of SARS-CoV-2, laboratory animals develop immunity inducing cross-protection. After Roborovski dwarf hamsters had been immunized with the att-Wuhan-like strain, the researchers infected them with Alpha and Beta virus variants circulating at the time of the study. The single-dose intranasal immunization was sufficient to establish protection in the hamsters infected not only with the parent strain of the virus, but also with the unrelated Alpha and Beta variants [52]. The same researchers confirmed the development of immunity providing cross-protection in Syrian hamsters immunized with the sCPD9 att-strain and then infected with the Delta strain [57].
Yoshida et al. immunized hamsters intranasally with attenuated and parent SARS-CoV-2 B.1.1 (Wuhan-like) strains and then infected them with the Omicron variant (the BA.1 lineage). In the nasal washes and lung homogenates from the immunized hamsters, the infectious titer was measured at the limit of detection and did not show any significant differences regardless of the strain selected for the immunization, thus being indicative of the development of cross-protection against phylogenetically distant strains, among other strains [17].
However, in hamsters immunized with wild-type strains of Beta and Omicron variants BA.1 and subsequently infected with Wuhan-like, Beta, Delta and Omicron BA.1 strains, Ma et al. revealed low cross-activity of antibodies in the hamsters infected with heterologous strains. At the same time, the hamsters previously infected with Omicron and then infected with heterologous Omicron strains demonstrated a booster effect on the production of neutralizing antibodies [58]. Thus, in the authors’ opinion, the two-dose infection with heterologous strains induces a strong antiviral immune response, which should be taken into consideration when designing vaccination schedules.
Conclusion
Most of the studies performed using animal models for COVID-19 demonstrated safety and high efficacy of intranasal immunization of animals with att-variants of SARS-CoV-2 regardless of the chosen strategy of virus attenuation. The pilot studies confirmed the assumption about protective activity of att-strains of SARS-CoV-2 toward heterologous antigenic variants of the virus. The animal model-based findings reported by researchers regarding the virulence and immunogenicity of candidate vaccine strains can hardly be extrapolated to humans without clinical studies. Nevertheless, despite this limitation, the currently gained experience indicates promising prospects in the development of a highly effective LAV against COVID-19.
Funding source. The study was supported by the Russian Science Foundation grant No. 23-25-00146, https://rscf.ru/project/23-25-00146/
Conflict of interest. The authors declare no apparent or potential conflicts of interest related to the publication of this article.
1 URL: https://covid19.who.int
2 URL: https://codagenix.com
About the authors
Ekaterina R. Korchevaya
I. Mechnikov Research Institute for Vaccines and Sera
Author for correspondence.
Email: c.korchevaya@gmail.com
ORCID iD: 0000-0002-6417-3301
junior researcher, Laboratory of applied virology, Department of Virology
Russian Federation, MoscowAnastasiia V. Gracheva
I. Mechnikov Research Institute for Vaccines and Sera
Email: c.korchevaya@gmail.com
ORCID iD: 0000-0001-8428-4482
researcher, Laboratory of applied virology, Department of Virology
Russian Federation, MoscowIlya N. Dyakov
I. Mechnikov Research Institute for Vaccines and Sera
Email: c.korchevaya@gmail.com
ORCID iD: 0000-0001-5384-9866
PhD (Biol.), Head of Laboratory of biosynthesis of immunoglobulins, Department of Virology
Russian Federation, MoscowVitaly V. Zverev
I. Mechnikov Research Institute for Vaccines and Sera
Email: c.korchevaya@gmail.com
ORCID iD: 0000-0001-5808-2246
D. Sci. (Biol.), Prof., Academician of RAS, scientific director
Russian Federation, MoscowEvgeny B. Faizuloev
I. Mechnikov Research Institute for Vaccines and Sera; Russian Medical Academy of Continuous Professional Education
Email: c.korchevaya@gmail.com
ORCID iD: 0000-0001-7385-5083
PhD (Biol.), Head, Laboratory of applied virology
Russian Federation, Moscow; MoscowReferences
- Teo S.P. Review of COVID-19 mRNA vaccines: BNT162b2 and mRNA-1273. J. Pharm. Pract. 2022;35(6):947–51. DOI: DOI: https://doi.org/10.1177/08971900211009650
- Wang H., Zhang Y., Huang B., et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182(3):713–21.e9. DOI: https://doi.org/10.1016/j.cell.2020.06.008
- Wu Z., Hu Y., Xu M., et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine (CoronaVac) in healthy adults aged 60 years and older: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21(6):803–12. DOI: https://doi.org/10.1016/S1473-3099(20)30987-7
- Al-Sheboul S.A., Brown B., Shboul Y., et al. An immunological review of SARS-CoV-2 infection and vaccine serology: innate and adaptive responses to mRNA, adenovirus, inactivated and protein subunit vaccines. Vaccines (Basel). 2022;11(1):51. DOI: https://doi.org/10.3390/vaccines11010051
- Khoshnood S., Arshadi M., Akrami S., et al. An overview on inactivated and live-attenuated SARS-CoV-2 vaccines. J. Clin. Lab. Anal. 2022;36:e24418. DOI: https://doi.org/10.1002/jcla.24418
- Bowen J.E., Addetia A., Dang H.V., et al. Omicron spike function and neutralizing activity elicited by a comprehensive panel of vaccines. Science. 2022;377(6608):890–4. DOI: https://doi.org/10.1126/science.abq0203
- Dejnirattisai W., Huo J., Zhou D., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467–84.e15. DOI: https://doi.org/10.1016/j.cell.2021.12.046
- Feikin D.R., Higdon M.M., Abu-Raddad L.J., et al. Duration of effectiveness of vaccines against SARS-CoV-2 infection and COVID-19 disease: results of a systematic review and meta-regression. Lancet. 2022;399(10328):924–44. DOI: https://doi.org/10.1016/S0140-6736(22)00152-0
- Chen J.M. Should the world collaborate imminently to develop neglected live-attenuated vaccines for COVID-19? J. Med. Virol. 2022;94(1):82–7. DOI: https://doi.org/10.1002/jmv.27335
- Goławski M., Lewandowski P., Jabłońska I., Delijewski M. The reassessed potential of SARS-CoV-2 attenuation for COVID-19 vaccine development – a systematic review. Viruses. 2022;14(5):991. DOI: https://doi.org/10.3390/v14050991
- Minor P.D. Live attenuated vaccines: Historical successes and current challenges. Virology. 2015;479-480:379–92. DOI: https://doi.org/10.1016/j.virol.2015.03.032
- Zimmerman L.A., Reef S.E., Orenstein W.A. Rubella vaccine-a tale of appropriate caution and remarkable success. JAMA Pediatr. 2018;172(1):95–6. DOI: https://doi.org/10.1001/jamapediatrics.2017.4178
- Li X.F., Cui Z., Fan H., et al. A highly immunogenic live-attenuated vaccine candidate prevents SARS-CoV-2 infection and transmission in hamsters. Innovation (Camb.). 2022;3(2):100221. DOI: https://doi.org/10.1016/j.xinn.2022.100221
- Maassab H.F., DeBorde D.C. Development and characterization of cold-adapted viruses for use as live virus vaccines. Vaccine. 1985;3(5):355–69. DOI: https://doi.org/10.1016/0264-410x(85)90124-0
- Trimpert J., Dietert K., Firsching T.C., et al. Development of safe and highly protective live-attenuated SARS-CoV-2 vaccine candidates by genome recoding. Cell Rep. 2021;36(5):109493. DOI: https://doi.org/10.1016/j.celrep.2021.109493
- Wang Y., Yang C., Song Y., et al. Scalable live-attenuated SARS-CoV-2 vaccine candidate demonstrates preclinical safety and efficacy. Proc. Natl Acad. Sci. USA. 2021;118(29):e2102775118. DOI: https://doi.org/10.1073/pnas.2102775118
- Yoshida A., Okamura S., Torii S., et al. Versatile live-attenuated SARS-CoV-2 vaccine platform applicable to variants induces protective immunity. iScience. 2022;25(11):105412. DOI: https://doi.org/10.1016/j.isci.2022.105412
- Liu Y., Zhang X., Liu J., et al. A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions. Nat. Commun. 2022;13(1):4337. DOI: https://doi.org/10.1038/s41467-022-31930-z
- Liu S., Stauft C.B., Selvaraj P., et al. Intranasal delivery of a rationally attenuated SARS-CoV-2 is immunogenic and protective in Syrian hamsters. Nat. Commun. 2022;13(1):6792. DOI: https://doi.org/10.1038/s41467-022-34571-4
- Ye Z.W., Ong C.P., Tang K., et al. Intranasal administration of a single dose of a candidate live attenuated vaccine derived from an NSP16-deficient SARS-CoV-2 strain confers sterilizing immunity in animals. Cell Mol. Immunol. 2022;19(5):588–601. DOI: https://doi.org/10.1038/s41423-022-00855-4
- Seo S.H., Jang Y. Cold-adapted live attenuated SARS-Cov-2 vaccine completely protects human ACE2 transgenic mice from SARS-Cov-2 infection. Vaccines (Basel). 2020;8(4):584. DOI: https://doi.org/10.3390/vaccines8040584
- Abdoli M., Shafaati M., Ghamsari L.K., Abdoli A. Intranasal administration of cold adapted live-attenuated SARS-CoV-2 candidate vaccine confers protection against SARS-CoV-2. Virus Res. 2022;319:198857.
- Faizuloev E., Gracheva A., Korchevaya E., et al. Cold-adapted SARS-CoV-2 variants with different temperature sensitivity exhibit an attenuated phenotype and confer protective immunity. Vaccine. 2022;41(4):892–902. DOI: https://doi.org/10.1016/j.vaccine.2022.12.019
- Xu J., Liu M., Niu X., et al. The cold-adapted, temperature-sensitive SARS-CoV-2 strain TS11 is attenuated in syrian hamsters and a candidate attenuated vaccine. Viruses. 2023;15(1):95. DOI: https://doi.org/10.3390/v15010095
- Hilleman M.R., Buynak E.B., Weibel R.E., et al. Development and evaluation of the Moraten measles virus vaccine. JAMA. 1968;206(3):587–90.
- Plotkin S.A., Buser F. History of RA27/3 rubella vaccine. Rev. Infect. Dis. 1985;7(Suppl. 1):S77–8. DOI: https://doi.org/10.1093/clinids/7.supplement_1.s77
- Maassab H.F. Adaptation and growth characteristics of influenza virus at 25 degrees c. Nature. 1967;213(5076):612–4. DOI: https://doi.org/10.1038/213612a0
- Smorodintsen A.A., Alexandrova G.A., Chalkova O.U., Selivanov A.A. Experiences in the development of live vaccines against influenza and influenza-like respiratory infections. Ind. Med. Surg. 1965;34:53–64.
- Ghendon Y.Z., Polezhaev F.I., Lisovskaya K.V., et al. Recombinant cold-adapted attenuated influenza A vaccines for use in children: molecular genetic analysis of the cold-adapted donor and recombinants. Infect. Immun. 1984;44(3):730–3. DOI: https://doi.org/10.1128/iai.44.3.730-733.1984
- Murphy B.R., Coelingh K. Principles underlying the development and use of live attenuated cold-adapted influenza A and B virus vaccines. Viral Immunol. 2002;15(2):295–323. DOI: https://doi.org/10.1089/08828240260066242
- Maassab H.F., Bryant M.L. The development of live attenuated cold-adapted influenza virus vaccine for humans. Rev. Med. Virol. 1999;9(4):237–44. DOI: https://doi.org/10.1002/(sici)1099-1654(199910/12)9:4 < 237::aid-rmv252 > 3.0.co;2-g
- Файзулоев Е.Б., Корчевая Е.Р., Грачева А.В. и др. Биологическая характеристика холодоадаптированных вариантов коронавируса SARS-CoV-2. Журнал микробиологии, эпидемиологии и иммунобиологии. 2022;99(4):397–409. Faizuloev E.B., Korchevaya E.R., Gracheva A.V., et al. Biological characterization of cold-adapted SARS-CoV-2 variants. Journal of Microbiology, Epidemiology and Immunobiology. 2022;99(4):397–409. DOI: https://doi.org/10.36233/0372-9311-280. EDN: https://elibrary.ru/llgegh
- Грачева А.В., Корчевая Е.Р., Самойликов Р.В. и др. Маркеры аттенуации холодоадаптированных вариантов коронавируса SARS-CoV-2. Медицинский академический журнал. 2022; 22(2):79–88. Gracheva A.V., Korchevaya E.R., Samoilikov R.V., et al. Attenuation markers of cold-adapted SARS-CoV-2 variants. Medical Academic Journal. 2022;22(2):79–88. DOI: https://doi.org/10.17816/MAJ108725
- Flavell R.A., Sabo D.L., Bandle E.F., Weissmann C. Site-directed mutagenesis: effect of an extracistronic mutation on the in vitro propagation of bacteriophage Qbeta RNA. Proc. Natl Acad. Sci. USA. 1975;72(1):367–71. DOI: https://doi.org/10.1073/pnas.72.1.367
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc. Natl Acad. Sci. USA. 1978;75(5):2170–4. DOI: https://doi.org/10.1073/pnas.75.5.2170
- Edelheit O., Hanukoglu A., Hanukoglu I. Simple and efficient site-directed mutagenesis using two single-primer reactions in parallel to generate mutants for protein structure-function studies. BMC Biotechnol. 2009;9:61. DOI: https://doi.org/10.1186/1472-6750-9-61
- Coleman J.R., Papamichail D., Skiena S., et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320(5884):1784–7. DOI: https://doi.org/10.1126/science.1155761
- Groenke N., Trimpert J., Merz S., et al. Mechanism of virus attenuation by codon pair deoptimization. Cell Rep. 2020;31(4):107586. DOI: https://doi.org/10.1016/j.celrep.2020.107586
- Zhang Z., Liu Q., Sun Y., et al. Live attenuated coronavirus vaccines deficient in N7-methyltransferase activity induce both humoral and cellular immune responses in mice. Emerg. Microbes Infect. 2021;10(1):1626–37. DOI: https://doi.org/10.1080/22221751.2021.1964385
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–27. DOI: https://doi.org/10.1038/s41586-020-2798-3
- Miteva D., Peshevska-Sekulovska M., Snegarova V., et al. Mucosal COVID-19 vaccines: Risks, benefits and control of the pandemic. World J. Virol. 2022;11(5):221–36. DOI: https://doi.org/10.5501/wjv.v11.i5.221
- Mateus J., Grifoni A., Tarke A., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370(6512):89–94. DOI: https://doi.org/10.1126/science.abd3871
- Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–501.e15. DOI: https://doi.org/10.1016/j.cell.2020.05.015
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184(4):861–80. DOI: https://doi.org/10.1016/j.cell.2021.01.007
- Tarke A., Sidney J., Kidd C.K., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2(2):100204. DOI: https://doi.org/10.1016/j.xcrm.2021.100204
- Geers D., Shamier M.C., Bogers S., et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021;6(59):eabj1750. DOI: https://doi.org/10.1126/sciimmunol.abj1750
- Nian X., Zhang J., Huang S., et al. Development of nasal vaccines and the associated challenges. Pharmaceutics. 2022;14(10):1983. DOI: https://doi.org/10.3390/pharmaceutics14101983
- Alu A., Chen L., Lei H., et al. Intranasal COVID-19 vaccines: From bench to bed. EBioMedicine. 2022;76:103841. DOI: https://doi.org/10.1016/j.ebiom.2022.103841
- Di Pietrantonj C., Rivetti A., Marchione P., et al. Vaccines for measles, mumps, rubella, and varicella in children. Cochrane Database Syst. Rev. 2021;11(11):CD004407. DOI: https://doi.org/10.1002/14651858.CD004407.pub5
- Ma S.J., Li X., Xiong Y.Q., et al. Combination measles- mumps-rubella-varicella vaccine in healthy children: a systematic review and meta-analysis of immunogenicity and safety. Medicine (Baltimore). 2015;94(44):e1721. DOI: https://doi.org/10.1097/MD.0000000000001721
- Mehla R., Kokate P., Bhosale S.R., et al. A live attenuated COVID-19 candidate vaccine for children: protection against SARS-CoV-2 challenge in hamsters. Vaccines (Basel). 2023; 11(2):255. DOI: https://doi.org/10.3390/vaccines11020255
- Trimpert J., Adler J.M., Eschke K., et al. Live attenuated virus vaccine protects against SARS-CoV-2 variants of concern B.1.1.7 (Alpha) and B.1.351 (Beta). Sci. Adv. 2021;7(49):eabk0172. DOI: https://doi.org/10.1126/sciadv.abk0172
- Deng L., Li P., Zhang X., et al. Risk of SARS-CoV-2 reinfection: a systematic review and meta-analysis. Sci. Rep. 2022;12(1): 20763. DOI: https://doi.org/10.1038/s41598-022-24220-7
- Mao Y., Wang W., Ma J., et al. Reinfection rates among patients previously infected by SARS-CoV-2: systematic review and meta-analysis. Chin. Med. J. (Engl.). 2021;135(2):145–52. DOI: https://doi.org/10.1097/CM9.0000000000001892
- Altarawneh H.N., Chemaitelly H., Hasan M.R., et al. Protection against the omicron variant from previous SARS-CoV-2 infection. N. Engl. J. Med. 2022;386(13):1288–90. DOI: https://doi.org/10.1056/NEJMc2200133
- Chemaitelly H., Nagelkerke N., Ayoub H.H., et al. Duration of immune protection of SARS-CoV-2 natural infection against reinfection. J. Travel Med. 2022;29(8):taac109. DOI: https://doi.org/10.1093/jtm/taac109
- Nouailles G., Adler J.M., Pennitz P., et al. Live-attenuated vaccine sCPD9 elicits superior mucosal and systemic immunity to SARS-CoV-2 variants in hamsters. Nat. Microbiol. 2023;8(5):860–74. DOI: https://doi.org/10.1038/s41564-023-01352-8
- Ma J., Liu X., Zhou M., et al. A heterologous challenge rescues the attenuated immunogenicity of SARS-CoV-2 omicron BA.1 variant in Syrian hamster model. J. Virol. 2023;97(2):e0168422. DOI: https://doi.org/10.1128/jvi.01684-22
Supplementary files