Analysis of the gene structure of antiphage systems of non-toxigenic strains of Vibrio cholerae O1 biovar El Tor
- Authors: Zadnova S.P.1, Plekhanov N.A.1, Sergutin D.A.1, Cheldyshova N.B.1, Fedorov A.V.1, Krasnov Y.M.1
-
Affiliations:
- Research Anti-Plague Institute “Microbe”
- Issue: Vol 102, No 4 (2025)
- Pages: 456-464
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18749
- DOI: https://doi.org/10.36233/0372-9311-640
- EDN: https://elibrary.ru/XNHHRX
- ID: 18749
Cite item
Abstract
Introduction. The presence and structure of antiphage systems that contribute to the resistance of cholera vibrios to lytic phages in non-toxigenic strains of Vibrio cholerae O1 biovar El Tor isolated in the Russian Federation and neighboring countries has not been studied.
The aim of the study is the detection and analysis of antiphage systems of non-toxigenic strains of V. cholerae O1 biovar El Tor.
Materials and methods. The study involved 126 non-toxigenic (ctxAB–tcpA+ and ctxAB–tcpA–) strains of V. cholerae O1 El Tor isolated from 1972 to 2018. DNA sequencing was performed on the MGI DNBSEQ-G50 platform. For bioinformatics analysis, the following programs were used: fastp v. 0.23, unicycler v. 0.4.7, Blast 2.16.0, MEGA X, CRISPRCasty-per and CRISPRCasFinder.
Results. Phage-inducible islands of the PLE, BREX and DISARM systems were not detected in the genome of the studied strains. It was found that 80% of ctxAB–tcpA+ strains contain the type I restriction-modification system, while this system was not detected in ctxAB–tcpA– isolates. The genes of the CBASS system were detected in single strains of both groups. In the genome of 35 (32%) studied ctxAB–tcpA– strains isolated in different regions of the Russian Federation and neighboring countries, the presence of the CRISPR–Cas system of class 1 types I (subtypes I-E, I-F, I-C) and III (subtype III-B) was established. The number of spacers in this system varied from 0 to 80 and their sequence was homologous to the protospacer regions of DNA of lytic and temperate phages, transposons, plasmids of V. cholerae, representatives of the genus Vibrio and unrelated bacteria. The presence in a number of strains of spacers homologous to the genetic material of the phage circulating in endemic territories may indicate the imported nature of these strains.
Conclusion. The heterogeneity of the studied non-toxigenic strains of V. cholerae O1 El Tor in the presence of antiphage systems was revealed, which expands the information on their genetic organization. In their genome, restriction-modification systems of type I (ctxAB–tcpA+), CBASS (ctxAB–tcpA+ and ctxAB–tcpA–) and CRISPR–Cas class 1 types I (subtypes I-E, I-F, I-C) and III (ctxAB–tcpA–) were identified. The detection of several types and subtypes of the CRISPR–Cas system in the genome of a number of ctxAB–tcpA strains may indicate its repeated acquisition through horizontal transfer.
Full Text
Introduction
Every year, during monitoring studies of Russian water bodies for cholera, non-toxigenic strains of Vibrio cholerae O1 serogroup El Tor biovar are isolated, lacking the ctxAB genes (ctxAB–) that encode the biosynthesis of cholera toxin (the main virulence factor of the cholera pathogen). Sometimes strains containing the tcpA gene (ctxAВ–tcpA+), which is responsible for the production of the main subunit of toxin-coregulated pili of adhesion (colonization factor), are isolated. Both ctxAB–tcpA– and ctxAB–tcpA+ strains do not cause cholera, but they could be the cause of acute intestinal infections. It has been established that non-toxigenic strains of V. cholerae O1 El Tor can persist in environmental objects in Russia for a long time, forming clonal complexes, and are also imported from cholera-endemic countries [1–7]. To identify the mechanisms of long-term survival of non-toxigenic strains in the external environment, as well as their ability to cause acute intestinal infections, the molecular genetic characteristics of these strains are being actively studied, various kinds of typing are being performed, and phylogenetic analysis is being conducted. As a result, the presence of genes encoding additional virulence and persistence factors has been identified in the genomes of non-toxigenic strains, which can also act as toxic substances. These include loci that provide motility, are responsible for the biosynthesis of additional toxins, a thermolabile hemolysin, proteases, neuraminidase, type 6 and type 3 secretion system proteins, mannose-sensitive adhesion pili, as well as regulatory proteins that control the transcription of virulence and persistence genes [3, 5, 8]. However, the reasons for the long-term survival of non-toxigenic V. cholerae O1 El Tor strains in the external environment are not fully understood.
It should be noted that when in the water of open bodies of water, non-toxigenic V. cholerae O1 El Tor vibrios can be attacked by bacteriophages (phages), which are also present in this environment. It has been shown that cholera phages play an important role in the genetic diversity of V. cholerae strains [9]. The co-existence of V. cholerae and phages and the need for both to survive drive the evolution of both V. cholerae and phages [9, 10]. Bacteria acquire various mechanisms of resistance to phages through horizontal gene transfer, while phages, in turn, very quickly develop resistance to many bacterial defense systems. It has been found that genes encoding resistance to phages can make up more than 10% of a bacterial genome [11].
A significant number of antiphage systems located on mobile genetic elements have been identified in toxigenic V. cholerae O1 El Tor strains. This includes a gene cluster encoding a type I restriction-modification system (vc1764–vc1769) located on the VPI-2 pathogenicity island, whose action is based on the activity of two enzymes: a restriction endonuclease (vc1765) and a methyltransferase (vc1769) [12]. The VSP-I pathogenicity island contains the CBASS (cyclic-oligonucleotide-based antiphage signaling system) antiphage signaling system, which includes an operon of 4 genes: dncV, capV, cap2, cap3 [13, 14]. The ICE SXT element contains BREX (Bacteriophage exclusion) and DISARM (Defence Islands System Associated with Restriction-Modification) systems [10, 15, 16].
A significant role in protection against the most common phage in the endemic area, ICP1, is played by Phage inducible chromosomal island-like elements (PLE), of which there are currently 10 types [17–19]. However, toxigenic El Tor vibrios lack the adaptive defense system CRISPR–Cas, which was identified in the genome of non-toxigenic V. cholerae O1 El Tor strains circulating in endemic areas [20]. This system includes clustered regularly interspaced short palindromic repeats (CRISPR), spacers (sequences of foreign origin), and cas genes encoding proteins with various functions [13]. Currently, the CRISPR–Cas system is classified into 2 classes, 6 types and 33 subtypes based on its mechanism of action, the structure of the CRISPR spacers, and the presence of cas genes.
In a Class 1 system (types I, III, IV), a multi-component complex consisting of several Cas proteins bound to crRNA interacts with the target. Class 2 systems (types II, V, and VI) contain only one protein (Cas9, Cas12, or Cas13) that performs all the functions of a multi-protein effector complex.
The classification into types is based on the structure of the effector complexes, with systems of the same type typically including a specific protein unique to that type of system. The types, in turn, are divided into subtypes, which differ in the structure of the CRISPR locus and, in certain cases, in the presence of Cas proteins. It is a fact that many Type I systems contain the cas3 gene, which encodes the Cas3 helicase-nuclease. Through its helicase activity, it unwinds the foreign DNA double helix, and with the involvement of its nuclease domain, it fragments the foreign genetic material. In type III systems, the Cas10 protein exhibits nuclease activity. All types include the Cas1 and Cas2 proteins, which form a complex and are responsible for the adaptation stage, i.e., the insertion of a new spacer into the CRISPR array [20, 21].
Despite active research on V. cholerae antiphage systems, their presence in the genomes of non-toxigenic V. cholerae O1 El Tor strains isolated in Russia and neighboring countries has not been studied. Considering the above, the aim of the study was to identify and analyze the antiphage systems of non-toxigenic strains of V. cholerae O1 El Tor biovar.
Materials and methods
We studied the nucleotide sequences of the complete genomes of 126 non-toxigenic V. cholerae O1 El Tor strains isolated in Russia and neighboring countries from 1972 to 2018. The nucleotide sequences of 30 strains (12 ctxA–tcpA+ and 18 ctxA–tcpA–) were obtained from the NCBI GenBank database, and 96 strains (3 ctxA–tcpA+ and 93 ctxA–tcpA–) were sequenced in this study. For sequencing, the strains were obtained from the State Collection of Pathogenic Bacteria of the Russian Anti-Plague Institute "Microbe" of Rospotrebnadzor, where they were stored in a lyophilized state.
Genomic DNA was prepared according to the manufacturer's protocol from a sodium merthiolate-treated bacterial suspension using the AxyPrep Bacterial Genomic DNA Miniprep Kit (Axygen Biosciences).
Sequencing was performed on the MGI DNBSEQ-G50 platform (MGI). Libraries were prepared according to a standard protocol using the DNBSEQ-G50RS (FCLPE150) and MGI EasyFastPCR-FREEFS Library PrepSet (MGI) kits. Quality control of the obtained reads was performed using the fastpv.0.23 program, and contigs were assembled using unicyclerv.0.4.7.
The Blast 2.16.0 algorithm (http://blast.ncbi) and the MEGA X program (or BioEditV. 7.0.9.0) were used for bioinformatics analysis. CRISPR–Cas systems and spacers were identified using the CRISPRCastyper (https://github.com/Russel88/CRISPRCasTyper) and CRISPRCasFinder (https://github.com/dcouvin/CRISPRCasFinder) programs.
Results
When studying the nucleotide sequences of complete genomes of non-toxigenic strains, phage-inducible PLE islands were not found in the genomes of either ctxAB–tcpA– or ctxAB–tcpA+ strains. The BREX and DISARM systems were also not detected, as these strains lack the ICE SXT elements.
Analysis of type I restriction-modification system genes (vc1765, vc1769) among 15 ctxAB–tcpA+ strains revealed 12 isolates that matched the toxigenic V. cholerae N16961 O1 El Tor reference strain for these genes (Table). In ctxAB–tcpA– strains, type I restriction-modification system genes were not detected.
Characteristics of certain non-toxigenic V. cholerae O1 El Tor strains and the structure of their anti-phage genes
Strain | Location, year | Source of isolation | Gene structure | Type of CRISPR–Cas systems | Amount of spacers | |
vc1765/ vc1769 | vc0179 (dncV)/ vc0178 (capV) | |||||
Non-toxigenic ctxАВ–tcpА+-strains | ||||||
М1395LQBY01 | Russia, Astrakhan, 1981 | Environment | – | – | – | – |
56MWRD01 | Ukraine, Mariupol, 1995 | Infected patient | int | int | – | – |
866MWRF01 | Ukraine, Yalta, 1996 | Environment | int | – | – | – |
85NEDU01 | Ukraine, Berdyansk, 1999 | Infected patient | int | – | – | – |
Р18778NIFI01 | Russia, Rostov-on-Don, 2005 | Infected patient | – | – | – | – |
М1434*, М1436* | Russia, Kalmykia, 2006 | Environment | int | – | – | – |
М1501LRAE01 | Russia, Kalmykia, 2011 | Infected patient | – | – | – | – |
М-1504VTLN01 | Russia, Kalmykia, 2011, 2012 | Environment | int | – | – | – |
М-1518LQZR01 | Russia, Kalmykia, 2011, 2012 | Environment | int | – | – | – |
М-1524LQZS01, М1528* | Russia, Kalmykia, 2013 | Environment | int | – | – | – |
2613PYCA01, 2687PYCB01 | Russia, Kalmykia, 2015 | Environment | int | – | – | – |
124PYCD01 | Russia, Kalmykia, 2017 | Environment | int | – | – | – |
Non-toxigenic ctxАВ–tcpА–-strain | ||||||
М-988LQBX01 | Turkmenistan, 1972 | Environment | – | – | I-F mini | Undetermined |
М-658* | Russia, Ufa, 1976 | Environment | – | – | I-F mini | 1 |
М-659* | Russia, Salavat, 1976 | Environment | – | – | I-E | 56 |
М-1114* | Russia, Saransk, 1977 | Infected patient | – | – | I-E I-F mini | 13 0 |
М-1115* | Russia, Saransk, 1977 | Infected patient | – | – | I-E I-F mini | 13 1 |
М-1394* | Russia, Kaspiysk, 1979 | Environment | – | – | I-E | 44 |
Strain | Location, year | Source of isolation | Gene structure | Type of CRISPR–Cas systems | Amount of spacers |
|
|
|
| vc1765/ vc1769 | vc0179 (dncV)/ vc0178 (capV) |
|
|
М-1222* | Russia, Astrakhan, 1985 | Environment | – | – | I-F mini | 10 |
М-1320* | Russia, Saratov, 1998 | Environment | – | – | III-B | 24 |
617NCTY01 | Ukraine, 1999 | Infected patient | – | – | I-F** I-E III-В | 26 80 25 |
М1337NEEB01 | Russia, Astrakhan, 2000 | Infected patient | – | – | I-E** | 1 |
М-1388* | Russia, Saratov, 2001 | Environment | – | – | I-E I-F mini | 13 1 |
М-1389* | Russia, Saratov, 2001 | Environment | – | – | I-E I-F mini | 13 1 |
М-1411* | Russia, Kalmykia, 2002 | Infected patient | – | – | I-F mini | 1 |
М-1413* | Russia, Kalmykia, 2002 | Environment | – | – | I-C I-F mini | 40 2 |
М-1426* | Russia, Perm Krai, 2003 | Environment | – | – | I-F mini | 0 |
М-1428* | Russia, Astrakhan, 2003 | Environment | – | – | I-E | 44 |
М-1431* | Russia, Kalmykia, 2005 | Environment | – | – | I-E | 60 |
Р-18748NIFH01 | Russia, Sochi, 2004 | Infected patient | – | A1003G (S335G) /int | – | – |
102NDXO01 | Ukraine, 2006 | Infected patient | – | A1003G (S335G) /int | I-F** | 49 |
М-1441* | Russia, Kalmykia, 2007 | Environment | – | – | I-E | 48 |
М-1443* | Russia, Kalmykia, 2007 | Environment | – | – | I-E | 58 |
М-1444* | Russia, Kalmykia, 2007 | Environment | – | – | I-E | 76 |
М-1447* | Russia, Kalmykia, 2009 | Environment | – | – | I-F | 15 |
М-1450* | Russia, Kalmykia, 2009 | Environment | – | – | I-F mini | 1 |
М1457VTLH01 | Russia, Kalmykia, 2009 | Environment | – | A1003G (S335G) /int | I-F | 2 |
М-1460* | Russia, Tatarstan, 2010 | Environment | – | – | I-E I-F mini | 14 1 |
2403NEDV01 | Ukraine, 2011 | Infected patient | – | – | I-E I-F mini | 12 0 |
М-1486* | Russia, Tatarstan, 2011 | Environment | – | – | I-E I-F mini | 29 1 |
М-1487* | Russia, Tatarstan, 2011 | Environment | – | – | I-E I-F mini | 15 0 |
М1516VTZY01 | Russia, Kalmykia, 2012 | Environment | – | – | I-F I-C | 58 54 |
М1517VTZZ01 | Russia, Kalmykia, 2012 | Environment | – | – | I-E | 23 |
М-1525* | Russia, Kalmykia, 2012 | Environment | – | – | I-F mini | 10 |
М1526VUAA01 | Russia, Kalmykia, 2012 | Environment | – | – | I-F I-C | 58 54 |
29VUAB01 | Russia, Kalmykia, 2013 | Environment | – | – | I-E | 7 |
М-1543* | Russia, Kalmykia, 2017 | Environment | – | – | I-E I-F mini | 29 1 |
136VTLK01 | Russia, Kalmykia, 2018 | Environment | – | – | I-E I-F mini | 8 3 |
Note. The strain superscript indicates the NCBI GenBank accession code. *Strains sequenced in this study. "–" — gene(s) not detected; int — the gene structure corresponds to the V. cholerae N16961 O1 El Tor reference strain. **The presence of the CRISPR–Cas system was previously established [20].
The presence of CBASS system genes (dncV, capV) was established in 4 strains. In the ctxAB–tcpA+ strain of V. cholerae 56, the gene data structure did not differ from that of the V. cholerae N16961 O1 El Tor reference strain. In ctxAB–tcpA– V. cholerae strains P-18748, 102, and M-1457, the capV gene was intact, and identical non-synonymous point mutations were identified in the dncV gene, leading to amino acid substitutions whose impact on the functional role of the DncV protein is unknown. In the genomes of other strains, the system mentioned was absent (Table).
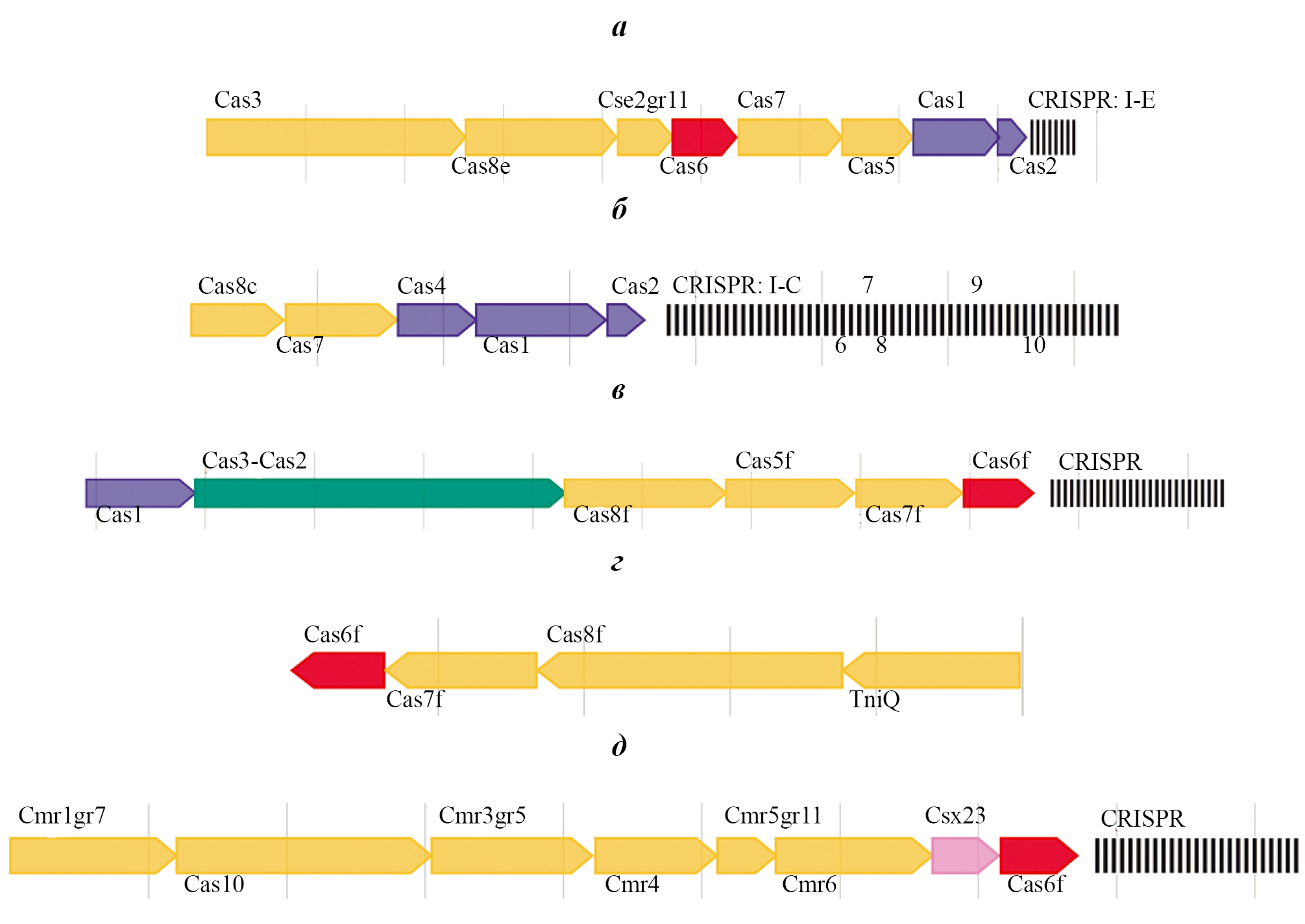
Next, the presence of the CRISPR–Cas system was investigated. This system was not found in the genomes of ctxAB–tcpA+ strains, while among the ctxAB–tcpA– strains, 35 isolates were identified that have a Class 1 Type 2 CRISPR–Cas system — Types I and III. Type I was represented by three subtypes: I-E, I-F, I-C, while Type III was represented by one (III-B). The largest number of strains (20 isolates) were found to have a subtype I-E CRISPR–Cas system, of which 10 isolates had only this subtype, while the others had additional systems. Thus, strain V. cholerae 617, in addition to I-E, had 2 more systems (Table). It should be noted that the I-E system structure was canonical in all strains and consisted of 8 cas genes (cas3, cas8e, cse2, cas6, cas7, cas5, cas1, cas2) (Figure, a).
Three strains — V. cholerae M1413, M1516, M1526 — belonged to subtype I-C (Table, Figure, b). Five strains (V. cholerae 617, 102, M1457, M1516, M1526) contained a complete subtype I-F CRISPR–Cas system with 6 cas genes (cas1, cas3, cas8f, cas5, cas7, cas6f: Table, Figure, c).
Structure of CRISPR–Cas systems in the studied ctxAB–tcpA– V. cholerae O1 El Tor strains.
a — canonical subtype I-E, present in V. cholerae strain 29; b — subtype I-C of V. cholerae strain M1526; c — subtype I-F of V. cholerae strain 617; d — subtype I-F mini of V. cholerae strain M1426; e — subtype III-B of V. cholerae strain M1320. The spacers (CRISPR) are marked by black vertical lines on the right.
A truncated I-F system (cas6f, cas7f, cas8f), consisting of 3 cas genes, which we have designated I-F mini (Figure, d), was identified in a number of strains. It should be noted that this system lacks the cas3 gene. Two strains, V. cholerae M1320 and 617, were type III, subtype III-B (cmr1, cas10, cmr3, cmr4, cmr5, cmr6, csx23, cas6f; Table, Figure, e)
Non-toxigenic ctxA–tcpA– strains with the CRISPR–Cas system are quite widespread in Russia. These strains were isolated in different years in the Republics of Tatarstan, Dagestan, Bashkortostan, Mordovia, Kalmykia, as well as in the Astrakhan, Saratov regions and the Perm Krai (Table).
The next stage of the study was dedicated to identifying spacers. They were present in almost all systems, with their number varying from 1 to 80, and the largest number of spacers was found in system I-E. The exception was 4 strains containing the I-F mini system, in which spacers were absent (Table). Due to the poor quality of the whole-genome nucleotide sequence presented in GenBank, it was not possible to reliably identify the spacers in strain M-988. The identified spacers were homologous to the protospacer sequences of a large number of lytic and temperate phages, as well as plasmids and the V. cholerae transposon (phages: O395, VPUSM 8, K139, K491, K571, K575, VcP032, Kappa, Rostov 7, X29, phi 2, JSF1, JSF2, JSF4, JSF5, JSF6, JSF13, JSF14, JSF17, VMJ710, Rostov M3, CP-T1, 24, vB_VchM-138, vB_VchM_VP-3213, Ch457, E8498, fs1, fs2, Vb_VaM_Valp1, ICP1, VRU, VP24-2_Ke, VMJ710, VcP032, VEJphi, VSK, VSKK, ND1-fs1, KSF-1phi, VGJphi, 1.178). O.J. L 286. 45. E12, 1.028. O.J. L 286. 45. B6, 1.159. O.J. L 261. 46. F12, Martha 12B12, Jenny 12G5, vB_Vipa26, vB_Vipa10, vB_Vipa4291, vB_Vipa71, vB_VpS_PG07, Zoerhiza.4_15, 13VV501A, 6E35-1b, D481, D483, D485, D491s, D527, VaK; plasmids: HDW18, pSA7G1, pSA7G2; transposon Tn7005). Furthermore, spacers homologous to the nucleotide sequences of phages and plasmids from Vibrio genus (V. alginolyticus, V. vulnificus, V. parahaemolyticus, V. fluvialis, V. furnissii, V. nigripulchritudo, V. metschnikovii), as well as from unrelated bacterial species (Klebsiella spp., Escherichia spp., Salmonella spp., Shigella, Shewanella algae, Xanthomonas, Stenotrophomonas) were identified. It is worth noting the presence of spacers in certain strains (V. cholerae 102, M1428, M1431, M1443, M1444, M1457, M1460, M1486) that are identical to DNA sequences of phage ICP1 (the most common bacteriophage in endemic areas), isolated in different years in the Democratic Republic of Congo and Bangladesh.
Thus, the following antiphage systems were identified in the genomes of the studied non-toxigenic V. cholerae O1 El Tor strains: type I restriction-modification (ctxAB–tcpA+), CBASS (ctxAB–tcpA+ and ctxAB–tcpA–) and class 1 CRISPR–Cas (ctxAB–tcpA–).
Discussion
Given that non-toxigenic V. cholerae O1 El Tor strains circulate in open water bodies, which are also habitats for various cholera phages, it was expected that a large number of antiphage systems would be found in their genomes. However, these strains lack the PLE, BREX, and DISARM antiphage islands of the system. Type I restriction-modification systems, which were found in the genome of ctxAB–tcpA+ strains, were not detected in ctxAB–tcpA– strains. CBASS system genes are present in a few strains from both groups. Meanwhile, 35 ctxAB–tcpA– strains studied, isolated from aquatic environments in various regions of Russia and neighboring countries, contain a Class 1 CRISPR–Cas system of types I and III, which is absent in ctxAB–tcpA+ isolates. However, subtypes I-C and III-B were detected in only a few strains, while 57% of the strains have the canonical type of the type I-E system. Our data confirm previously obtained information from the analysis of strains circulating in the endemic area regarding the widespread distribution of the I-E system among non-toxigenic V. cholerae strains [20]. A number of authors suggest that the stable maintenance of the I-E system structure is due to its location on the GI-24 genomic island, and its transfer to other strains occurs only as part of this mobile genetic element [20, 22]. Certain strains also included a complete I-F system, which is part of the newly discovered VPI-6 (Vibrio Pathogenicity Island), capable, similar to GI-24, of being entirely excised from the chromosome and transferred to other cells [23]. At the same time, most strains had the I-F mini system, which has the tniQ gene responsible for transposase production located next to the cas gene loci. According to literature data, such systems are associated with the Tn7 transposon [20]. Given the absence of the cas3 gene encoding a helicase-nuclease in this system, it can be assumed that I-F mini is non-functional.
Since new spacers are primarily inserted into the 5' regions of the system, CRISPR represents a chronological record of the bacterium's interaction with mobile genetic elements. In this regard, the presence of spacers homologous to the genetic material of phage ICP1 circulating in endemic areas may indicate the imported nature of these strains. It is also evident that by protecting V. cholerae from predation by cholera phages, as well as phages of other bacteria, the CRISPR–Cas system increases the survival of non-toxigenic strains in the external environment. Perhaps its presence is one of the mechanisms for the long-term circulation of non-toxigenic strains in open water bodies.
Thus, the studied non-toxigenic ctxAB–tcpA+ and ctxAB–tcpA– V. cholerae O1 El Tor strains isolated in Russia and neighboring countries lack a number of mobile genetic elements with anti-phage loci (PLE islands, ICE SXT elements with BREX and DISARM systems). At the same time, a type I restriction-modification system was identified in the genome of ctxAB–tcpA+ strains, and a CBASS (ctxAB–tcpA+ and ctxAB–tcpA–) and a class 1 CRISPR–Cas system (ctxAB–tcpA–) were identified in one strain.
Conclusion
The research conducted has established the heterogeneity of the studied non-toxigenic V. cholerae O1 El Tor strains circulating in Russia and neighboring countries in terms of the presence of antiphage systems located on mobile genetic elements, which expands our knowledge of their genetic organization. It was found that 80% of the studied ctxAB–tcpA+ strains contain a type I restriction–modification system, which was not detected in ctxAB–tcpA– strains. Single strains (1 ctxAB–tcpA+ and 3 ctxAB–tcpA–) have a CBASS system, which is intact in the ctxAB–tcpA+ isolate and corresponds to the toxigenic reference strain V. cholerae N16961 O1 El Tor. A class 1 CRISPR–Cas system of type I (subtypes I-E, I-F, I-F mini, I-C) and type III (subtype III-B), which is absent in ctxAB–tcpA+ strains, was identified in the genome of 32% of the ctxAB-tcpA- strains studied. The most common (57%) is the canonical system of subtype I-E. The presence of multiple types and subtypes of the CRISPR–Cas system in the genome of several strains may indicate its repeated acquisition by these isolates through horizontal transfer. Analysis of spacers in the CRISPR cassette allows for the identification of non-toxigenic V. cholerae O1 El Tor strains imported from cholera-endemic areas.
About the authors
Svetlana P. Zadnova
Research Anti-Plague Institute “Microbe”
Email: svetlanazadnova@mail.ru
ORCID iD: 0000-0003-4366-0562
Dr. Sci. (Biol.), leading researcher, Head, Laboratory of pathogenic vibrios
Russian Federation, SaratovNikita A. Plekhanov
Research Anti-Plague Institute “Microbe”
Email: muscari.sp@icloud.com
ORCID iD: 0000-0002-2355-7018
Cand. Sci. (Biol.), senior researcher, Laboratory of pathogenic vibrios
Russian Federation, SaratovDanil A. Sergutin
Research Anti-Plague Institute “Microbe”
Author for correspondence.
Email: sergutin322@mail.ru
ORCID iD: 0009-0003-7525-9682
Laboratory assistant, Laboratory of pathogenic vibrios
Russian Federation, SaratovNadezhda B. Cheldyshova
Research Anti-Plague Institute “Microbe”
Email: rusrapi@microbe.ru
ORCID iD: 0000-0001-5759-3765
Cand. Sci. (Med.), senior researcher, Laboratory of pathogenic vibrios
Russian Federation, SaratovAndrey V. Fedorov
Research Anti-Plague Institute “Microbe”
Email: rusrapi@microbe.ru
ORCID iD: 0000-0001-7190-4427
Junior researcher, Laboratory of genomic and proteomic analysis
Russian Federation, SaratovYaroslav M. Krasnov
Research Anti-Plague Institute “Microbe”
Email: rusrapi@microbe.ru
ORCID iD: 0000-0002-4909-2394
Cand. Sci. (Chem.), Head, Laboratory of genomic and proteomic analysis
Russian Federation, SaratovReferences
- Смирнова Н.И., Кульшань Т.А., Баранихина Е.Ю. и др. Структура генома и происхождение нетоксигенных штаммов Vibrio cholerae биовара Эль Тор с различной эпидемиологической значимостью. Генетика. 2016;52(9):1029–41. DOI: https://doi.org/10.7868/S0016675816060126 EDN: https://elibrary.ru/wlnekj Smirnova N.I., Kul’shan’ T.A., Baranikhina E.Y., et al. Genome structure and origin of nontoxigenic strains of Vibrio cholerae of El Tor biovar with different epidemiological significance. Genetics. 2016; 52(9): 1029–41. DOI: https://doi.org/10.1134/S1022795416060120 EDN: https://elibrary.ru/xfheir
- Кругликов В.Д., Левченко Д.А., Титова С.В. и др. Холерные вибрионы в водоёмах Российской Федерации. Гигиена и санитария. 2019;98(4):393–9. Kruglikov V.D., Levchenko D.A., Titova S.V., et al. Vibrio cholerae in the waters of the Russian Federation. Gigiena i Sanitaria (Hygiene and Sanitation, Russian journal). 2019;98(4):393–9. EDN: https://elibrary.ru/dfqoan
- Левченко Д.А., Кругликов В.Д., Гаевская Н.Е. и др. Фено- и генотипические особенности нетоксигенных штаммов холерных вибрионов различного происхождения, изолированных на территории России. Проблемы особо опасных инфекций. 2020;(3):89–96. Levchenkо D.А., Kruglikov V.D., Gaevskaya N.E., et al. Pheno- and genotypical features of non-toxigenic strains of cholera vibrios of different origins, isolated in the territory of Russia. Problems of Particularly Dangerous Infections. 2020;(3):89–96. DOI: https://doi.org/10.21055/0370-1069-2020-3-89-96 EDN: https://elibrary.ru/rvjkhr
- Миронова Л.В., Бочалгин Н.О., Гладких А.С. и др. Филогенетическое положение и особенности структуры геномов ctxAB–tcpA+ Vibrio cholerae из поверхностных водоемов на неэндемичной по холере территории. Проблемы особо опасных инфекций. 2020;(1):115–23. Mironova L.V., Bochalgin N.O., Gladkikh A.S., et al. Phylogenetic affinity and genome structure features of ctxAB–tcpA+ Vibrio cholerae from the surface water bodies in the territory that is non-endemic as regards cholera. Problems of Particularly Dangerous Infections. 2020;(1):115–23. DOI: https://doi.org/10.21055/0370-1069-2020-1-115-123 EDN: https://elibrary.ru/uubigv
- Монахова Е.В., Носков А.К., Кругликов В.Д. и др. Генотипическая характеристика клональных комплексов CTX–VPI+ Vibrio cholerae О1, обнаруживаемых в водоемах Ростовской области. Проблемы особо опасных инфекций. 2023;(3):99–107. Monakhova E.V., Noskov A.K., Kruglikov V.D., et al. Genotypic characteristics of CTX–VPI+ clonal complexes of Vibrio cholerae O1 found in water bodies of the Rostov region. Problems of Particularly Dangerous Infections. 2023;(3):99–107. DOI: https://doi.org/10.21055/0370-1069-2023-3-99-107
- Носков А.К., Кругликов В.Д., Москвитина Э.А. и др. Холера: анализ и оценка эпидемиологической обстановки в мире и России. Прогноз на 2023 г. Проблемы особо опасных инфекций. 2023;(1):56–66. Noskov A.K., Kruglikov V.D., Moskvitina E.A., et al. Cholera: analysis and assessment of epidemiological situation around the world and in Russia (2013–2022). Forecast for 2023. Problems of Particularly Dangerous Infections. 2023;(1):56–66. DOI: https://doi.org/10.21055/0370-1069-2023-1-56-66 EDN: https://elibrary.ru/hzasbo
- Попова А.Ю., Носков А.К., Ежлова Е.Б. и др. Эпидемиологическая ситуация по холере в Российской Федерации в 2023 г. и прогноз на 2024 г. Проблемы особо опасных инфекций. 2024;(1):76–88. Popova A.Yu., Noskov A.K., Ezhlova E.B., et al. Epidemiological situation on cholera in the Russian Federation in 2023 and forecast for 2024. Problems of Particularly Dangerous Infections. 2024;(1):76–88. DOI: https://doi.org/10.21055/0370-1069-2024-1-76-88 EDN: https://elibrary.ru/ipvmuo
- Агафонова Е.Ю., Смирнова Н.И., Альхова Ж.В. и др. Нетоксигенные штаммы Vibrio cholerae биовара Эль Тор, выделенные на территории России: молекулярно-генетические особенности и патогенные свойства. Журнал микробиологии, эпидемиологии и иммунобиологии. 2019;96(2):13–24. Agafonova E.Yu., Smirnova N.I., Alkhova Zh.V., et al. On-toxigenic strains of Vibrio cholerae biovar El Tor, isolated in the territory of Russia: molecular-genetic peculiarities and pathogenic properties. Journal of microbiology, epidemiology and immunobiology. 2019;96(2):13–24. DOI: https://doi.org/10.36233/0372-9311-2019-2-13-24 EDN: https://elibrary.ru/yijsem
- Faruque S.M., Mekalanos J.J. Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence. 2012;3(7):556–65. DOI: https://doi.org/10.4161/viru.22351
- Angermeyer A., Hays S.G., Nguyen M.H.T., et al. Evolutionary sweeps of subviral parasites and their phage host bring unique parasite variants and disappearance of a phage CRISPR-Cas system. mBio. 2022;13(1):e03088-21. DOI: https://doi.org/10.1128/mbio.03088-21
- Tumban E., ed. Bacteriophages. Methods and Protocols. New York: Humana Press; 2024.
- Jermyn W.S., Boyd E.F. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology. 2002;148(Pt. 11):3681–93. DOI: https://doi.org/10.1099/00221287-148-11-3681
- Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8(5):317–27. DOI: https://doi.org/10.1038/nrmicro2315
- Brenzinger S., Airoldi M., Ogunleye A.J., et al. The Vibrio cholerae CBASS phage defence system modulates resistance and killing by antifolate antibiotics. Nat. Microbiol. 2024;9(1):251–62. DOI: https://doi.org/10.1038/s41564-023-01556-y
- LeGault K.N., Hays S.G., Angermeyer A., et al. Temporal shifts in antibiotic resistance elements govern phage-pathogen conflicts. Science. 2021;373(6554):eabg2166. DOI: https://doi.org/10.1126/science.abg2166
- Заднова С.П., Плеханов Н.А., Спирина А.Ю., Челдышова Н.Б. Анализ антифаговых систем в штаммах Vibrio cholerae O1 биовара Эль Тор. Здоровье населения и среда обитания – ЗНиСО. 2023;31(11):94–100. Zadnova S.P., Plekhanov N.A., Spirina A.Yu., Cheldyshova N.B. Analysis of Antiphage Systems in Vibrio cholerae O1 El Tor Biotype Strains. Public Health and Life Environment – PH&LE. 2023;31(11):94–100. DOI: https://doi.org/10.35627/2219-5238/2023-31-11-94-100 EDN: https://elibrary.ru/miocic
- O’Hara B.J., Barth Z.K., McKitterick A.C., Seed K.D. A highly specific phage defense system is a conserved feature of the Vibrio cholerae mobilome. PLoS Genet. 2017;13(6):e1006838. DOI: https://doi.org/10.1371/journal.pgen.1006838
- McKitterick A.C., Seed K.D. Anti-phage islands force their target phage to directly mediate island excision and spread. Nat. Commun. 2018;9(1):2348. DOI: https://doi.org/10.1038/s41467-018-04786-5
- Barth Z.K., Silvas T.V., Angermeyer A., Seed K.D. Genome replication dynamics of a bacteriophage and its satellite reveal strategies for parasitism and viral restriction. Nucleic Acids Res. 2020;48(1): 249–63. DOI: https://doi.org/10.1093/nar/gkz1005
- McDonald N.D., Regmi A., Morreale D.P., et al. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genomics. 2019;20(1):105. DOI: https://doi.org/10.1186/s12864-019-5439-1
- Makarova K.S., Wolf Y.I., Iranzo J., et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020;18(2):67–83. DOI: https://doi.org/10.1038/s41579-019-0299-x
- Chakraborty S., Waise T.M., Hassan F., et al. Assessment of the evolutionary origin and possibility of CRISPR-Cas (CASS) mediated RNA interference pathway in Vibrio cholerae O395. In Silico Biol. 2009;9(4):245–54.
- Carpenter M.R., Kalburge S.S., Borowski J.D., et al. CRISPR-Cas and contact-dependent secretion systems present on excisable pathogenicity islands with conserved recombination modules. J. Bacteriol. 2017;199(10):e00842-16. DOI: https://doi.org/10.1128/jb.00842-16
Supplementary files