The influence of an innovative antibacterial drug of the thiadiazinone class on the virulence factors of bacteria of the phylum Pseudomonadota, which chronically infect patients with cystic fibrosis
- Authors: Voronina O.L.1, Koroleva E.A.1, Kunda M.S.1, Ryzhova N.N.1, Aksenova E.I.1, Kapotina L.N.1, Nelubina S.A.1, Lazareva A.V.2, Zigangirova N.A.1
-
Affiliations:
- N.F. Gamaleya National Research Center for Epidemiology and Microbiology
- National Medical Research Center for Children's Health
- Issue: Vol 101, No 2 (2024)
- Pages: 173-183
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/18537
- DOI: https://doi.org/10.36233/0372-9311-499
- EDN: https://elibrary.ru/pmwtsz
- ID: 18537
Cite item
Abstract
Introduction. Infections of the lower respiratory tract by bacteria of the Pseudomonadota phylum: Pseudomonas aeruginosa, Burkholderia spp., Achromobacter spp. are critical to the quality and life expectancy of patients with cystic fibrosis (CF). When the infection is chronic, eradication of bacteria with existing antibacterial drugs is practically impossible. To explore alternative drugs, trials are needed on bacteria isolated from CF patients and characterized using genomic approaches.
The objective of our study was a comparative analysis of virulence factors of 6 isolates of bacteria of the Pseudomonadota phylum and testing the efficacy of the innovative drug Fluorothiazinone (FT) in suppressing the pathogenicity of bacteria in vitro.
Materials and methods. Isolates of A. ruhlandii ST36, A. xylosoxidans ST555, B. cepacia ST2140, B. gladioli ST2141, P. aeruginosa ST859 and ST198 were examined using whole-genome sequencing and bioinformatics analysis to search for resistance and virulence determinants. The FT drug was tested for its effect on bacteria in in vitro experiments on cytotoxicity on HeLa cells, motility and biofilm formation.
Results. Genomic studies have confirmed the arsenal of resistance determinants, especially the efflux systems of bacteria isolated from patients with CF, and the diversity of virulence factors, among which we identified factors in the categories of motility, signals of quorum-sensing systems, secretion systems, exotoxins, as the most essential for the adaptation of bacteria to conditions of the lower respiratory tract. In vitro tests of the FT drug showed its effectiveness in suppressing cytotoxicity (2.6–4.0 times), motility (2.0–3.6 times) and the process of biofilm formation (2.0–7.7 times).
Conclusion. For the first time, the effectiveness of the innovative antibacterial drug Fluorothiazinone has been shown against bacteria of the Pseudomonadota phylum, isolated from chronically infected patients with CF, with the described potential of virulence factors.
Full Text
Introduction
Cystic fibrosis (CF) is one of the most common autosomal recessive diseases in which mutations in the gene for the transmembrane regulator of the chlorine channel cause impaired mucociliary clearance and the development of chronic colonization of the respiratory tract by bacteria of the Pseudomonadota phylum. Microorganisms of this phylum – Pseudomonas aeruginosa, Burkholderia spp. and Achromobacter spp. – are characterized by high natural resistance to antimicrobial drugs. Despite the use of aggressive antibiotic therapy, a progressive decline in lung function and early mortality are observed in CF patients with age. According to the latest edition of the Russian CF patient registry, the proportion of patients chronically infected with the listed bacteria is 33.6% for P. aeruginosa, 7.6% for Achromobacter spp. and 5.5% for Burkholderia spp. [1].
Penetrating the lower respiratory tract by aspiration, bacteria move along the surface of epitheliocytes using flagella and pili/fimbriae; these same structures serve as adhesins when attaching to cells to initiate biofilm formation [2]. Afterwards, several migration pathways of bacteria of these genera are possible: translocation through intercellular contacts and transepithelial migration [3]. In the latter case, invasion is first accomplished through the secretion systems of types 3 and 6 (T3SS, T6SS). The bacteria that have emerged from the epitheliocytes then cross the basal membrane and reach the connective tissue cells. On this pathway, they are protected by exotoxins that affect collagen and interfere with its antimicrobial activity [4]. P. aeruginosa, Achromobacter spp. and Burkholderia spp. can carry out invasion into macrophages, neutrophils and dendritic cells, thus being able to be protected from external influences in 4 types of eukaryotic cells. Bacteria not only survive inside cells, but can also disrupt their normal functioning, leading to pyroptosis/apoptosis or necrosis [5]. Cell death, bacterial escape and further multiplication cause an inflammatory response that damages lung tissue [3]. All the above mentioned stages in the life cycle of pathogens are coordinated by Quorum-Sensing (QS) signaling [6]. Survival mechanisms help these bacteria to compete with each other and with other representatives of the lung microbiome, so microbial diversity in such infections becomes minimal [7].
The eradication of such successful pathogens requires new approaches, one of which was used in the development of an innovative antibacterial drug of the thiadiazinone class (fluorothiazinone, FT) at the N.F. Gamaleya National Research Center for Epidemiology and Microbiology. T3SS effectors [8], as well as, presumably, highly conserved ATPases of the flagellar apparatus and T3SS, became a target for the effect of FT, which suppresses the pathogenicity of bacteria but does not kill them [8, 9], which ensures the absence of resistance development to such a drug. The efficacy of FT in vitro and in animal models was shown against a number of Gram-negative bacteria: Salmonella enterica, P. aeruginosa, Escherichia coli, Acinetobacter baumannii and Klebsiella pneumoniae [10]. The drug inhibited the obligate intracellular pathogen Chlamydia trachomatis [11], proving the possibility of FT penetration into eukaryotic cells.
Earlier, in the study of P. aeruginosa isolates from sputum and tracheal aspirate of CF patients, we showed that the cytotoxicity testing conditions selected for cultures isolated in nosocomial infections [9] were optimal only for isolates of genotypes ST235 and ST313 [12], characteristic of nosocomial P. aeruginosa belonging to the ExoU-lineage, named after the effector T3SS [13]. The other isolates were characterized by slow growth in vitro due to changes in cell physiology during chronic lung infection. Adaptation of experimental conditions to the peculiarities of Pseudomonadota of CF patients was one of the objectives of the study.
Considering the diversity of bacterial virulence factors used in adaptation and persistence in the respiratory tract of CF patients, we studied the genomic characteristics of isolates selected to evaluate the effect of FT and compared factors in the categories: motility, QS signaling, secretion systems, and exotoxins in selected representatives of P. aeruginosa, Burkholderia spp. and Achromobacter spp.
The objectives of our study were to comparatively analyze the virulence factors of 6 isolates of Pseudomonadota phylum bacteria infecting the lower respiratory tracts of CF patients and to test the efficacy of an innovative FT drug in suppressing the pathogenicity of bacteria in vitro.
Materials and methods
Materials
Six cultures of Pseudomonadota phylum bacteria were isolated from the sputum of chronically infected CF patients (Table 1). The study was conducted under the conditions of obtaining voluntary informed consent from patients or their legal representatives. The study protocol was approved by the Biomedical Ethics Committee of N.F. Gamaleya National Research Center for Epidemiology and Microbiology (protocol No. 59 of 08.09.2023).
FT is a novel antibacterial drug of thiadiazinone class C19H17F2N3O4S: N-(2,4-difluorophenyl)-4(3-ethoxy-4-hydroxybenzyl)-5-oxo-5,6-dihydro-4H-[1, 3, 4]-thiadiazine-2-carboxamide1. A stock solution of FT was prepared from the substance with a concentration of 5.0 mM in 0.3M CH3COONa, pH 7.0 ± 0.2.
Cytotoxicity was studied on HeLa cervical carcinoma cells (ATCC CCL2, 22603).
Bacteria cultivation
Bacteria were grown for 18 h at 37ºC in LB broth to a concentration of 109 microbial cells/mL (OD600).
Genome analysis
The protocol [14] was used for DNA extraction from isolates, supplemented by polysaccharide purification using CTAB (cetyltrimethylammonium bromide).
DNA libraries were prepared using the protocols Nextera DNA Flex Library Prep (Illumina) and KAPA HyperPlus Kit (Roche). Sequencing was performed on a NextSeq 500/550 instrument (Illumina) using a Mid Output 300 cycles cartridge.
Genomes were assembled using CLC Genomic Workbench v. 21.0.1 (Qiagen) and SPAdes v. 3.13.02. Rapid Annotations Subsystems Technology (RAST) [15] and NCBI Prokaryotic Genome Annotation Pipeline [16] were used for annotation. Results were deposited in GenBank (bioproject PRJNA561493) under the numbers shown in Table 1.
Table 1. Isolates of the Pseudomonadota phylum used in the study
Specie | Isolate | Accession number | Sequence type | Genome size, Mb | Genes | Protein-coding |
P. aeruginosa | GIMC5045:PA33P25 | JAVMRC000000000 | ST859 | 6,4 | 5973 | 5841 |
P. aeruginosa | GIMC5047:PA33P30 | JAVMRD000000000 | ST198 | 6,4 | 6023 | 5842 |
B. cepacia | SCCH90:Bcn202840 | JAQOTY000000000 | ST2140 | 8,4 | 7704 | 7512 |
B. gladioli | SCCH61:Bgd92-3601 | JAQOTZ000000000 | ST2141 | 8,2 | 9105 | 8385 |
A. ruhlandii | SCCH137:Ach2231057 | JAQZZN000000000 | ST36 | 6,3 | 5884 | 5679 |
A. xylosoxidans | SCCH131:Ach223717 | JAPZVF000000000 | ST555 | 6,4 | 5912 | 5806 |
Genomes were analyzed using BV-BRC resources3 [17]. Virulence factors were investigated using VFDB4 [18] and BlastKOALA5 [19]. Plasmids were searched using PlasmidFinder 2.16. CARD7 [20], BV-BRC [17] and BlastKOALA [19] were used to identify resistance determinants.
Investigation of the effect of FT in vitro
Bacterial cytotoxicity was determined according to the method [9], with modifications. A monolayer of HeLa cells grown in IMDM (Iscove's Modified Dulbecco's Medium) supplemented with 10% FBS (fetal bovine serum) and 2 mM L-glutamine in 96-well plates was washed and IMDM containing 1% FBS was added. HeLa cells were infected with bacterial cultures at an initial multiplicity of infection (MOI) of 5. Plates were incubated for 18 h in the presence of FT (60 μg/mL). 0.3M CH3COONa, pH 7.0 ± 0.2, was used as a control. Cells were precipitated by centrifugation for 20 min at 1500 rpm. In the supernatants, the activity of released lactate dehydrogenase (LDH) was determined using the CytoTox 96® Non-Radioactive Cytotoxicity Assay Kit (Promega) according to the manufacturer's protocol. The percentage of LDH release was calculated relative to uninfected control (0% LDH release) and HeLa cells lysed with Triton X-100 (100% LDH release).
The ability of FT to inhibit the swimming motility of isolates was evaluated on Petri dishes with 0.3% semi-liquid agar [21]. Bacterial cultures were incubated with FT (100 μg/mL) for 3 h at 37ºC, then 2 μL of the suspension was added to the thickness of semi-liquid agar containing FT (100 μg/mL) and incubated for 48 h at 37ºC. The degree of bacterial motility was determined by the diameter of radial migration in agar.
The following approach was used to study the effect of FT on bacterial biofilm formation. Static biofilms were formed on the abiotic surface according to the protocol [22] with changes in incubation conditions. FT (100 μg/mL) was added to overnight bacterial cultures at a concentration of 107 microbial cells/mL (OD600) and incubated in the wells of the plate for 48 h without changing the medium, then 125 μL of 0.1% crystal violet (CV) solution was added one at a time to stain the biofilms. The dye bound to the biofilms was extracted with 100 μl of 96% ethanol and OD540 was determined on a Multiskan EX instrument (Thermo Labsystems). Qualitative studies of biofilms were performed under a Nikon Eclipse 50i microscope (Nikon) at 20× magnification.
Each FT experiment was repeated 3 times.
Statistical processing of the analysis results and visualization were performed using Prism-GraphPad (GraphPad Software). The criterion of statistical reliability of the difference between the obtained data was considered to be the error value p < 0.05.
Results
Genome analysis
Six isolates of the Pseudomonadota phylum represented 3 genera. A. ruhlandii and A. xylosoxidans, selected for the study, were representatives of the genus most common in CF patients in Russia. The choice of B. cepacia and B. gladioli was determined by the emergence of new species of Burkholderia spp. infecting CF patients against the background of decreasing spread of B. cenocepacia of epidemic genotype ST709 [23]. P. aeruginosa of different genotypes belonging to the ExoS lineage, according to E.A. Ozer et al. [13], were taken into the study as more common in infections of CF patients compared to the ExoU phenotype [12, 24].
The genomes of the isolates were represented by one chromosome in Achromobacter and Pseudomonas and 3 chromosomes in Burkholderia. The 48 Kb conjugative plasmid was present only in the genome of A. ruhlandii (IncP1), but did not include resistance determinants. The size of Burkholderia genomes was one-third larger than Achromobacter and Pseudomonas genomes (Table 1). In all genomes, 92-98% of the identified genes encoded proteins.
When assessing the resistance potential of the studied isolates, the number of efflux systems encoded by genomes was noteworthy: 12 in P. aeruginosa, 16 in Achromobacter spp. each, 27 in B. cepacia, 38 in B. gladioli, which creates additional opportunities to counteract the applied antibiotic therapy.
Investigating the virulence factors of isolates, we focused on 4 main groups necessary for bacterial adaptation to lower respiratory tract conditions: secretion systems, motility, toxins, and QS signaling.
The secretion systems (Table 2) Sec, SRT, Tat, and T2SS are represented by all components in the genomes of the isolates. T3SS is found in all isolates except B. cepacia; T6SS is found in 5 isolates, and in A. ruhlandii it contains only genes of secreted substrate Hcp and inner membrane protein IcmF; finally, complete T1SS is found in B. gladioli, and in the other isolates it is represented only by outer membrane protein TolC.
Table 2. Classes of bacterial secretion systems represented in the genomes of the studied isolates
Classes of bacterial protein secretion systems | P. aeruginosa GIMC5045: PA33P25 | P. aeruginosa GIMC5047: PA33P30 | B. cepacia SCCH90: Bcn202840 | B. gladioli SCCH61: Bgd92-3601 | A. ruhlandii SCCH137: Ach2231057 | A. xylosoxidans SCCH131: Ach223717 |
Sec | + | + | + | + | + | + |
SRT | + | + | + | + | + | + |
Tat | + | + | + | + | + | + |
T1SS | + (TolC) | + (TolC) | + (TolC) | + (TolC, HlyB, HlyD) | + (TolC) | + (TolC) |
T2SS | + | + | + | + | + | + |
T3SS | + | + | + | + | + | |
T4SS | + VirD4 | + VirB5, VirB6 | ||||
T6SS | + | + | + | + | + Hcp, IcmF | + |
Note. VirD4 — ATPase; VirB5 — surface/pilus protein; VirB6, IcmF — inner membrane protein; Hcp — secreted substrate.
The main motility apparatus, the flagella apparatus, is present in the genomes of all isolates. The identified hfp pili/fimbriae differed in composition among the isolates. As shown in Table 3, pili responsible for twitching motility and chemosensory activity were found only in the genomes of P. aeruginosa. Type IV pili are present in pseudomonads and Burkholderia spp. but differ in the list of components, and in Achromobacter they are represented only by the prepilin pilD peptidase. Type IVb pili are present in all isolates. Chaperone-usher pili, which encode 2 different operons: fimACD and cupE1-6, are present in P. aeruginosa and B. cepacia with a complete set of components. In B. gladioli, the fimACD operon lacks a chaperone gene and the cupE operon is not detected. In Achromobacter genomes, the full cupE1-6 operon is present, while the fimACD operon lacks the gene encoding pilin.
Table 3. Pilus systems in the genomes of the studied isolates
Pilus systems | P. aeruginosa GIMC5045:PA33P25 | P. aeruginosa GIMC5047:PA33P30 | B. cepacia SCCH90:Bcn202840 | B. gladioli SCCH61:Bgd92-3601 | A. ruhlandii SCCH137:Ach2231057 | A. xylosoxidans SCCH131:Ach223717 |
Twitching motility pili | pilGHIJKRSTUKRS | pilGHIJKRSTUKRS | – | – | – | – |
Chemosensory pili | chpABCDE | chpABCDE | – | – | – | – |
Type IV pili | pilABCDFQPONMZVWXY1Y2E | pilABCDFQPNMZVWXY1Y2E | pilABCDEQW | pilABCDEW | pilD | pilD |
Type IVb pili | flp, cpaABCEF | flp, cpaABCEF | flp, cpaABCEF | flp, cpaABCEF | flp, cpaABCEF | flp, cpaABCEF |
Chaperone-Usher Pathway (CUP) pili | fimACD, cupE | fimACD, cupE | fimACD, cupE | fimAD | fimCD, cupE | fimCD, cupE |
Positive phototactic motility proteins | – | – | – | – | pixH | – |
Note. pilD — prepilin peptidase; pixH — response regulator.
The QS system as the most important means of bacterial communication is represented in the analyzed genomes in all its diversity. QS of AI-1 type (AutoInductor), whose signaling molecules are homoserinlactone derivatives, was found in the genomes of P. aeruginosa (2 each) and Burkholderia (1 each). An AI-1-regulated operon of rhamnolipid biosynthesis is also present in these genomes. Rhamnolipids are included in the QS system, are used by bacterial cells to reduce surface tension, and are important for motility, biofilm formation, and absorption of hydrophobic substrates [25].
The second system is named DSF from signaling molecules which are diffusible signaling factors. For Burkholderia spp. this BDSF is cys-2-dodecenoic acid. Another name for it is rpfF/R/B/G — by genes. Achromobacter genomes have 2 DSF systems each, B. cepacia has 1 DSF. The genomes of P. aeruginosa and B. gladioli have only rpfB genes.
The third system, PQS (pseudomonas quinolone signal), whose signaling molecule is 2-heptyl-3-hydroxyl-4-quinolone (C16H21NO2), in the complete set: pqsABCDHE, phnAB, is present only in P. aeruginosa. The genome of B. cepacia contains pqsE, a gene for a protein that responds to quinolone signaling, and phnAB, encoding an anthrenylate synthesis protein, a precursor of PQS [26]. Achromobacter and B. gladioli have only phnAB.
The toxins that the studied isolates are capable of producing can be divided into 4 groups. The first one is T3SS toxins acting inside the eukaryotic cell. In the genome of P. aeruginosa GIMC5045:PA33P25 they are represented by 5 genes: toxA, exoS, exoT, exoY, zot. The toxA gene encodes an ADP-ribosyltransferase. The zot gene is a homolog of cholera toxin acting on zonula occludens (the main one of the tight contacts proteins of the intestinal epithelium), the second isolate of P. aeruginosa has 4 genes of this group. The T3SS effector gene was also found in the genomes of Achromobacter — axoU. The second group contains genes of toxins that damage the membrane of eukaryotic cells. The genes of phospholipase C and hemolysin III are present in all genomes, the gene tlyC (pore-forming toxin) is absent in Achromobacter, and the genome of B. cepacia contains another gene of this group, tlh, encoding thermolabile hemolysin. In the third group of nonspecific toxins, only P. aeruginosa has hcnABC (hydrogen cyanide synthase) genes. The fourth group of toxins damaging the extracellular matrix is found only in B. cepacia and is represented by the colA gene of microbial collagenase.
Effect of FT in vitro
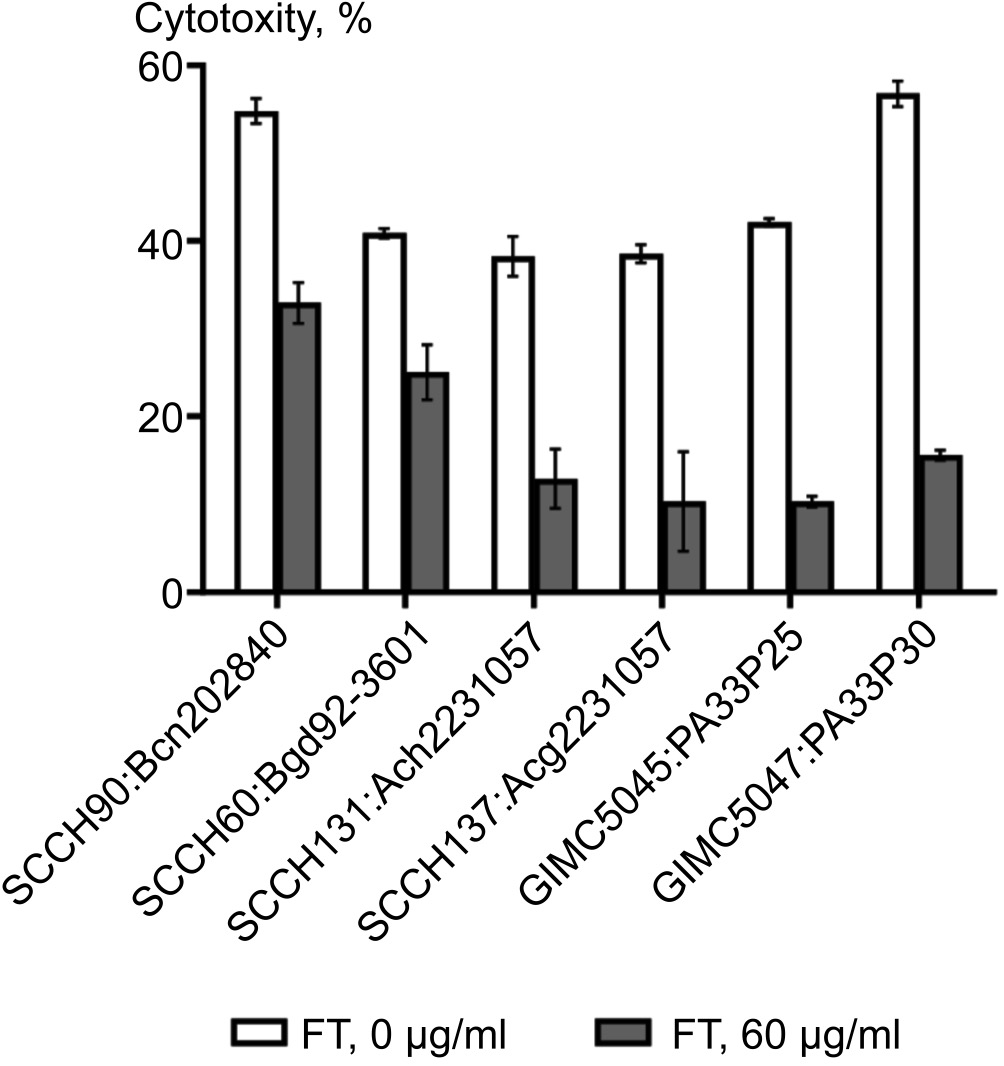
The effect of FT on the cytotoxicity of isolates from CF patients was investigated by selecting the time of contact of bacterial cells with HeLa cells, taking into account the slow growth of such bacteria in culture. While at a contact time of 4 h at doses of 10 and 50 MOI the cytotoxicity of isolates was 16–26 and 27–43%, respectively, after 20 h of contact the toxicity rose to 70-100%. A dose of 5 MOI and a contact time of 18 h were determined as optimal for studying the effect of FT. Under the influence of FT there was a decrease in cytotoxicity for all isolates: for P. aeruginosa isolates — by 3.6 and 4.0 times, for B. cepacia — by 2.6, for B. gladioli — by 3.2, for A. xylosoxidans — by 3.0, for A. ruhlandii — by 3.7 (Fig. 1).
Fig. 1. Effect of FT on the cytotoxicity of bacterial cells against HeLa cells.
Comparison of the swimming mobility of isolates showed a significant reduction of the bacterial movement zone under the influence of FT: for P. aeruginosa — 2.2 and 2.8 times, for B. cepacia — 2.7 times, for B. gladioli — 2.0 times, for A. xylosoxidans — 2.0 times, for A. ruhlandii — 3.6 times (Fig. 2). The mobility of P. aeruginosa isolates in the control was lower than other bacteria, however, the difference with samples incubated with FT was statistically significant (p < 0.05).
Fig. 2. Effect of FT on swimming mobility of bacterial cells.
The process of biofilm formation differed among isolates of 3 genera (Fig. 3). Burkholderia and Achromobacter were characterized by a fairly rapid development of dense biofilm structures over the entire well area, while for Pseudomonas biofilm formation was slower and was mainly concentrated at the edges of the well, where a dense ring was formed (Fig. 4). The effect of FT was strongest for B. cepacia. The biofilm biomass decreased 7.7-fold in the presence of the antibacterial agent. For the other isolates the decrease in biomass was lower but significant: for A. xylosoxidans and A. ruhlandii — 3-fold, for B. gladioli — 2.3-fold, for P. aeruginosa — 2.4-fold and 2.0-fold (Fig. 5).
Fig. 3. Сhanges in the diameter of bacterial swimming mobility zones.
Fig. 4. Effect of FT on the formation of bacterial biofilms. Microphotographs of the formed biofilm fragments (the densest fragments) are shown.
Fig. 5. Evaluation of biofilm biomass accumulation by the degree of crystal violet staining based on optical density (λ = 540 nm).
Discussion
Respiratory tract infections with P. aeruginosa, Achromobacter spp. and Burkholderia spp. are the most frequent and most dangerous for CF patients. Multiple natural resistance of the B. cepacia complex is already postulated and warnings about it are stated in the EUCAST antibiotic susceptibility testing guidelines8. The antibiogram for Achromobacter spp. is also a definite challenge for laboratories, as EUCAST thresholds are only given for 3 substances, even in the 2024 guidelines. For P. aeruginosa, the problem is that sensitive in vitro isolates cannot be eradicated with selected drugs. Our genomic studies have shown that the potential for resistance in P. aeruginosa and other bacteria studied may be the efflux systems present in genomes in large numbers. We should not forget about biofilms, the formation of which is successfully coordinated by QS signals, the presence and diversity of which we confirmed for all genomes studied. In vitro experiments showed that all 6 isolates formed dense biofilms. It is these structures that help bacteria avoid the effects of antibiotics in human lungs. However, the innovative FT drug was effective against all isolates tested.
Cytotoxicity of representatives of three genera is a serious problem for lung tissues of CF patients. The spectrum of exotoxins that can be produced by the studied isolates is quite wide. It should be noted that in the study of the proteomes of P. aeruginosa, Achromobacter spp. and Burkholderia spp. there are still many surprises and discoveries awaiting us, since at present it is possible to annotate a little more than half of the translation products that encode the genomes we have sequenced. The leader in annotation is P. aeruginosa (57.8%), with B. gladioli in last place at 38.2%. The databases of annotation resources lack, for example, the sequences of genes encoding the Achromobacter spp. T3SS effector, so we performed an additional search for the axoU gene, finding it in both Achromobacter genomes, named the hypothetical protein gene. For B. gladioli, the search for T3SS effectors is ongoing. S.K. Yadav et al. found the ortholog of the T3SS effector in B. gladioli strain NGJ1, showing the presence of a secretion signal at the N-terminus of a polypeptide annotated as a prophage protein, and demonstrated its calcium-dependent secretion mediated by T3SS [27]. The frame of such a protein is also present in the genome sequenced by us as part of the prophage. During in vitro experiments, all 6 isolates tested showed cytotoxicity against HeLa cells. The cytotoxicity of B. cepacia was at the level of the most effective P. aeruginosa isolate in the absence of T3SS, as shown by our genomic studies. It should be noted that the first publication mentioning the absence of T3SS in B. cepacia dates back to 2001. [28]. It is possible that another nanomachine, T6SS, is involved in the delivery of B. cepacia toxins, especially since in other bacteria T6SS and T3SS work in coordination [29].
Genomic studies have demonstrated an arsenal of factors responsible for motility of the studied bacteria of the Pseudomonadota phylum. The main one for swimming motility is flagella, the genes of which apparatus are present in all genomes. We observed this type of motility in the isolates tested, more pronounced in B. gladioli and Achromobacter under experimental conditions.
Conclusion
Thus, genomic studies and in vitro analysis of isolates allowed us to describe the virulence factors of 6 bacteria isolated from chronically infected patients and to demonstrate the possibility of their realization by all isolates for important processes in the development and chronicity of infection: cytotoxicity, motility and biofilm formation.
The innovative antibacterial agent FT inhibited the three processes in all isolates in vitro. The efficacy of FT against isolates from CF patients has been shown for the first time. These experiments will serve as a basis for further preclinical trials of the drug against a new nosology, including animal models. Ongoing comprehensive studies of FT itself have demonstrated accumulation of the drug administered intragastrically to rats in various animal organs, including the lungs [10]. Thus, the evidence base for the efficacy of FT in vivo and in vitro is constantly expanding, which gives hope that a new helper in the prevention and treatment of respiratory tract infections in CF patients may emerge.
1 Clinical studies: RCT No. 389 dated 08/03/2018 (completed); RKI 169 dated March 14, 2022 (ongoing). Application for registration with the Ministry of Health of the Russian Federation (incoming No. 4253550 dated May 30, 2023). Status under review.
2 St. Petersburg genome assembler, Russia,
URL: http://cab.spbu.ru/software/spades/
3 Bacterial and Viral Bioinformatics Resource Center,
URL: https://www.bv-brc.org
4 Virulence Factor Database, http://www.mgc.ac.cn/VFs
5 KEGG Orthology And Links Annotation,
URL: https://www.kegg.jp/blastkoala
6 URL: https://cge.food.dtu.dk/services/PlasmidFinder
7 Comprehensive Antibiotic Resistance Database,
URL: https://card.mcmaster.ca
8 Antimicrobial susceptibility testing of Burkholderia cepacia complex (BCC). 2013. URL: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/BCC_susceptibility_testing_130719.pdf
About the authors
Olga L. Voronina
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: olv550@gmail.com
ORCID iD: 0000-0001-7206-3594
Cand. Sci. (Biol.), Assistant Professor, Head, Laboratory of genome analysis
Russian Federation, MoscowEkaterina A. Koroleva
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: olv550@gmail.com
ORCID iD: 0000-0001-9702-2940
Cand. Sci. (Biol.), senior researcher, Laboratory of chlamydiosis
Russian Federation, MoscowMarina S. Kunda
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: olv550@gmail.com
ORCID iD: 0000-0003-1945-0397
Cand. Sci. (Biol.), senior researcher, Laboratory of genome analysis
Russian Federation, MoscowNatalia N. Ryzhova
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: olv550@gmail.com
ORCID iD: 0000-0001-5361-870X
Cand. Sci. (Biol.), senior researcher, Laboratory of genome analysis
Russian Federation, MoscowEkaterina I. Aksenova
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: olv550@gmail.com
ORCID iD: 0000-0003-2704-6730
Cand. Sci. (Biol.), senior researcher, Laboratory of genome analysis
Russian Federation, MoscowLidia N. Kapotina
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: olv550@gmail.com
ORCID iD: 0000-0003-0159-5053
Cand. Sci. (Biol.), senior researcher, Laboratory of chlamydiosis
Russian Federation, MoscowStanislava A. Nelubina
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Email: olv550@gmail.com
ORCID iD: 0000-0002-5157-1415
junior researcher, Laboratory of chlamydiosis
Russian Federation, MoscowAnna V. Lazareva
National Medical Research Center for Children's Health
Email: olv550@gmail.com
ORCID iD: 0000-0003-3896-2590
D. Sci. (Med.), chief researcher, Laboratory of molecular microbiology
Russian Federation, MoscowNailya A. Zigangirova
N.F. Gamaleya National Research Center for Epidemiology and Microbiology
Author for correspondence.
Email: olv550@gmail.com
ORCID iD: 0000-0003-3188-1608
D. Sci. (Biol.), Professor, chief researcher, Head, Laboratory of chlamydiosis
Russian Federation, MoscowReferences
- Регистр пациентов с муковисцидозом в Российской Федерации. 2021 год. СПб.;2023.
- Kazmierczak B.I., Mostov K., Engel J.N. Interaction of bacterial pathogens with polarized epithelium. Annu. Rev. Microbiol. 2001;55:407–35. doi: https://doi.org/10.1146/annurev.micro.55.1.407
- Saldías M.S., Valvano M.A. Interactions of Burkholderia cenocepacia and other Burkholderia cepacia complex bacteria with epithelial and phagocytic cells. Microbiology (Reading). 2009;155(Pt. 9):2809–17. doi: https://doi.org/10.1099/mic.0.031344-0
- Abdillahi S.M., Bober M., Nordin S., et al. Collagen VI is upregulated in COPD and serves both as an adhesive target and a bactericidal barrier for Moraxella catarrhalis. J. Innate Immun. 2015;7(5):506–17. DOI: https://doi.org/10.1159/000381213
- Li S.S., Saleh M., Xiang R.F., et al. Natural killer cells kill Burkholderia cepacia complex via a contact-dependent and cytolytic mechanism. Int. Immunol. 2019;31(6):385–96. doi: https://doi.org/10.1093/intimm/dxz016
- Subramani R., Jayaprakashvel M. Chapter 3. Bacterial quorum sensing: biofilm formation, survival behaviour and antibiotic resistance. In: Bramhachari P.V., ed. Implication of Quorum Sensing and Biofilm Formation in Medicine, Agriculture and Food Industry. Singapore;2019:21–37. doi: https://doi.org/10.1007/978-981-32-9409-7_3
- Pickrum A.M., DeLeon O., Dirck A., et al. Achromobacter xylosoxidans cellular pathology is correlated with activation of a type III secretion system. Infect. Immun. 2020;88(7):e00136-20. DOI: https://doi.org/10.1128/iai.00136-20
- Zigangirova N.A., Nesterenko L.N., Sheremet A.B., et al. Fluorothiazinon, a small-molecular inhibitor of T3SS, suppresses salmonella oral infection in mice. J. Antibiot. (Tokyo). 2021;74(4):244–54. doi: https://doi.org/10.1038/s41429-020-00396-w
- Sheremet A.B., Zigangirova N.A., Zayakin E.S., et al. Small molecule inhibitor of type three secretion system belonging to a class 2,4-disubstituted-4H-[1,3,4]-thiadiazine-5-ones improves survival and decreases bacterial loads in an airway Pseudomonas aeruginosa infection in mice. Biomed. Res. Int. 2018;2018:5810767. doi: https://doi.org/10.1155/2018/5810767
- Savitskii M.V., Moskaleva N.E., Brito A., et al. Dose proportional pharmacokinetics, organ distribution, bioavailability and excretion of the antivirulence drug Fluorothiazinon in rats and rabbits. J. Antibiot. (Tokyo). 2024. doi: https://doi.org/10.1038/s41429-024-00719-1
- Zigangirova N.A., Kost E.A., Didenko L.V., et al. A small-molecule compound belonging to a class of 2,4-disubstituted 1,3,4-thiadiazine-5-ones inhibits intracellular growth and persistence of Chlamydia trachomatis. J. Med. Microbiol. 2016;65(1):91–8. DOI: https://doi.org/10.1099/jmm.0.000189
- Воронина О.Л., Рыжова Н.Н., Кунда М.С. и др. Pseudomonas aeruginosa. Ассистенты и конкуренты в микробиоме инфицированных легких больных муковисцидозом. Медицинский вестник Северного Кавказа. 2020;15(2):186–91. Voronina O.L., Ryzhova N.N., Kunda M.S., et al. Pseudomonas aeruginosa. Assistants and competitors in the microbiome of infected of cystic fibrosis patients’ lungs. Medical News of North Caucasus. 2020;15(2):186–91. doi: https://doi.org/10.14300/mnnc.2020.15045 EDN: https://elibrary.ru/izwatj
- Ozer E.A., Nnah E., Didelot X., et al. The population structure of Pseudomonas aeruginosa is characterized by genetic isolation of exoU+ and exoS+ lineages. Genome Biol. Evol. 2019;11(1):1780–96. DOI: https://doi.org/10.1093/gbe/evz119
- Wilson K. Preparation of genomic DNA from bacteria. Curr. Protoc. Mol. Biol. 2001;Chapter 2:Unit 2.4. doi: 10.1002/0471142727.mb0204s56
- Brettin T., Davis J.J., Disz T., et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:8365. DOI: https://doi.org/10.1038/srep08365
- Li W., O'Neill K.R., Haft D.H., et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49(D1):D1020–8. DOI: https://doi.org/10.1093/nar/gkaa1105
- Olson R.D., Assaf R., Brettin T., et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023;51(D1):D678–89. doi: https://doi.org/10.1093/nar/gkac1003
- Chen L., Yang J., Yu J., et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33(Database issue):D325-8. DOI: https://doi.org/10.1093/nar/gki008
- Kanehisa M., Sato Y., Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016;428(4):726–31. DOI: https://doi.org/10.1016/j.jmb.2015.11.006
- Alcock B.P., Huynh W., Chalil R., et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023;51(D1):D690–9. doi: https://doi.org/10.1093/nar/gkac920
- Koroleva E.A., Soloveva A.V., Morgunova E.Y., et al. Fluorothiazinon inhibits the virulence factors of uropathogenic Escherichia coli involved in the development of urinary tract infection. J. Antibiot. (Tokyo). 2023;76(5):279–90. doi: https://doi.org/10.1038/s41429-023-00602-5
- Merritt J.H., Kadouri D.E., O'Toole G.A. Growing and analyzing static biofilms. Curr. Protoc. Microbiol. 2005;Chapter 1: Unit 1B.1. doi: https://doi.org/10.1002/9780471729259.mc01b01s00
- Воронина О.Л., Рыжова Н.Н., Кунда М.С. и др. Основные тенденции в изменении разнообразия буркхолдерий, инфицирующих российских больных муковисцидозом. Сибирское медицинское обозрение. 2019;(2):80–8. Voronina O.L., Ryzhova N.N., Kunda M.S., et al. Major tendencies in burkholderia diversity changes, infecting Russian patients with cystic fibrosis. Siberian Medical Review. 2019;(2):80–8. doi: https://doi.org/10.20333/2500136-2019-2-80-88 EDN: https://elibrary.ru/aqqxee
- Martínez-Alemán S., Bustamante A.E., Jimenez-Valdes R.J., et al. Pseudomonas aeruginosa isolates from cystic fibrosis patients induce neutrophil extracellular traps with different morphologies that could correlate with their disease severity. Int. J. Med. Microbiol. 2020;310(7):151451. doi: https://doi.org/10.1016/j.ijmm.2020.151451
- Wittgens A., Kovacic F., Müller M.M., et al. Novel insights into biosynthesis and uptake of rhamnolipids and their precursors. Appl. Microbiol. Biotechnol. 2017;101(7):2865–78. doi: https://doi.org/10.1007/s00253-016-8041-3
- Choi Y., Park H.Y., Park S.J., et al. Growth phase-differential quorum sensing regulation of anthranilate metabolism in Pseudomonas aeruginosa. Mol. Cells. 2011;32(1):57–65. doi: https://doi.org/10.1007/s10059-011-2322-6
- Yadav S.K., Das J., Kumar R., Jha G. Calcium regulates the mycophagous ability of Burkholderia gladioli strain NGJ1 in a type III secretion system-dependent manner. BMC Microbiol. 2020;20(1):216. doi: https://doi.org/10.1186/s12866-020-01897-2
- Parsons Y.N., Glendinning K.J., Thornton V., et al. A putative type III secretion gene cluster is widely distributed in the Burkholderia cepacia complex but absent from genomovar I. FEMS Microbiol. Lett. 2001;203(1):103–8. doi: https://doi.org/10.1111/j.1574-6968.2001.tb10827.x
- Le Goff M., Vastel M., Lebrun R., et al. Characterization of the Achromobacter xylosoxidans type VI secretion system and its implication in cystic fibrosis. Front. Cell. Infect. Microbiol. 2022;12:859181. doi: https://doi.org/10.3389/fcimb.2022.859181
Supplementary files