Nucleotide tetramers TCGA and CTAG: viral DNA and the genetic code (hypothesis)
- Authors: Filatov F.P.1,2
-
Affiliations:
- I. Mechnikov Research Institute of Vaccines and Sera
- National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
- Issue: Vol 99, No 4 (2022)
- Pages: 478-493
- Section: ORIGINAL RESEARCHES
- URL: https://microbiol.crie.ru/jour/article/view/1305
- DOI: https://doi.org/10.36233/0372-9311-275
- ID: 1305
Cite item
Abstract
Introduction. The published and our own data show that CTAG and, to a lesser extent, TCGA tetra-nucleotides have significantly lower concentrations in frequency profiles (FPs) of herpesvirus DNAs compared to other complete, bilaterally symmetrical tetra-nucleotides.
The aim of the study is to present a comparative analysis of CTAG and TCGA tetra-nucleotide FPs in viral DNAs.
Materials and methods. We have analyzed FPs and other characteristics of the two above tetramers in DNAs of at least one species of viruses of each genus (or each subfamily, if the classification into genera was not available), complying with the size limit requirements (minimum 100,000 base pairs) — a total of more than 200 species of viruses. The analysis was performed using the GenBank database.
Results. Two groups of characteristics of TCGA and CTAG tetramers have been described. One of them covers the results of the FP analysis for these tetranucleotides in viral DNAs and shows that DNAs with GC:AT > 2 are characterized by nCGn FP symmetries while these symmetries are frequently distorted in nTAn FP due to CTAG underrepresentation. The other group of tetramer characteristics demonstrates differences in their FPs in complete viral DNAs and in their genomes (a coding part, which can reach 80% in some studied viruses, thus making the analysis of their DNAs more significant than the analysis of DNAs of cellular live forms) and suggests that these tetramers may have participated in the origin of the universal genetic code.
Discussion. Assumedly, the genetic code started evolving amid C+G prevailing in "pre-code" DNA polymers; then the initial code forms evolved further to their final structure where TCGA and CTAG tetramers hold a central position, encapsulating the previous stages of this evolution. The nCGn FP symmetries typical of the "complete" DNA of Herpes simplex viruses disappear in the sequence of the second codon letters of the genome of these viruses, implying that their functions differ from functions of other letters and emphasizing the reasonableness of presenting the genetic code as a calligram where the second line is not symmetrical.
Full Text
INTRODUCTION
Earlier, we described the frequency of occurrence for bilaterally symmetrical, complete (consisting of 4 bases) tetra-nucleotides (TNs) in genomes of herpesviruses [1]. Having found that the frequency profiles (FPs) of two TNs — CTAG and, to a lesser extent, TCGA — of herpesvirus DNAs had significantly low concentrations, which was also supported by data of other published studies [2–4], we thoroughly analyzed other characteristics of the above TNs and extended the scope of the analysis beyond the boundaries of herpesviruses.
It is assumed that the CTAG function is associated with disruption of the optimal nucleic acid stem-loop structure, thus causing inhibition of DNA replication (the thermodynamic model). In addition, the CTAG sequence is more sensitive to chemical exposure [5][6]. TCGA owes its lower concentrations to its central dimer CpG, which is notable for its frequent methylation and mutations [7–10].
In this article, we have referred to multiple studies (though there are much more works addressing this subject) to show the diverse consequences of the discussed oligonucleotides in DNAs and genomes of living organisms [11]. Undesired inhibition of biological syntheses is offset by lower concentrations of both TNs in DNAs. Our primary attention was given to formal characteristics of both TNs, which, compared to the other, have biological functions, regardless of the governing functions and mechanisms. These characteristics demonstrate unexpected qualities, which will be explained here in the context of a provisional hypothesis.
The aim of the study is to present a comparative analysis of CTAG and TCGA tetra-nucleotide FPs in viral DNAs and genomic regions of these DNAs.
We analyzed the closest context of central pairs of CG and TA nucleotide tetramers, including TCGA and CTAG, in DNAs of viruses representing different taxonomic groups. This approach makes it easier to compare frequency profiles (FPs) of the CG dinucleotide and CTAG, approximating their sizes and treating them both as tetramers and dimers (especially when many researchers mention functionally similar, though to a significantly lesser extent, characteristics of the CTAG central dimer [12, 13]). The downside of this approach is that the densities of symmetric pairs of tetramers with the common function (TCGA and ACGT) show much fewer differences compared to the significantly different densities of symmetric pairs of tetramers having this function (CTAG) and hardly having it (GTAC).
MATERIALS AND METHODS
The analysis included physically unsegmented DNAs with full-length sequences available in GenBank1 as of 2021. The third limiting factor — the DNA size, which should be at least 100 kbp, as in case with Chargaff’s second parity rule [5][14][15], and not larger than 300–400 kbp. DNAs of the latter are typical of highly complex viruses and contain primarily A+T. The largest known viral RNAs — genomes of coronaviruses — are of no more than 32–35 kbp in size.
The above requirements were met by genomes of viruses only of two major realms of the Vira superkingdom: Duplodnaviria (the kingdom Heunggongvirae) and Varidnaviria (the kingdom Bamfordvirae). We analyzed DNAs of at least one species of viruses representing each genus (or subfamily, if it was not divided into genera); the total number of such genera was more than 200 (20 families). The studied viruses of the first realm belonged to phyla Uroviricota of the kingdom Heunggongvirae (the order Caudovirales) and Peploviricota of the same kingdom (the order Herpesvirales). The studied viruses of the other realm belonged to the phylum Nucleocytoviricota of classes Megaviricetes and Pokkesviricetes. We also analyzed DNAs of viruses without identified intermediate realms: 9 representatives of families Baculoviridae, Nudiviridae and the superfamily Nimaviridae as well as 6 representatives of unclassified archaeal viruses and 3 unclassified species of families Pytho- and Hytrosaviridae (Appendix).
GenBank programs were used as tools for the analysis.
Frequency distribution graphs for the studied TNs were built by searching variants of the closest context of central pairs nTAn and nCGn (CTAG) with a successively increasing molecular weight n [16, 17]:
[C→T]Y → [A→G]R,
where C, T — pyrimidines (Y); A, G — purines (R).
RESULTS
1. DENSITY OF NTAN AND NCGN OF THE MINIMUM CONCENTRATIONS IN VIRAL DNAS
The nTAn and nCGn density analysis showed that CTAGmin is the TN of the lowest concentration in DNAs of most (75 out of 128) of the studied representatives of phylum Uroviricota. The term “minimum concentration” refers to a complete self complementary tetramer with the density lower than the density of the contextually symmetrical tetramer in the general FP of the viral DNA. In our case, CTAGmin < GTAC and TCGAmin < ACGT.
DNA of any species of the phylum Uroviricota does not contain TCGA as the tetramer at the minimum concentration. In DNAs of viruses belonging to the phylum Nucleocytoviricota, unassigned viruses (Baculoviridae, Nudiviridae and Nimaviridae) and archaeal viruses, TCGAmin can be present, but its occurrence is rare and does not have any apparent relationship with classification groups.
One of the iridoviruses — the alpha-iridovirus as well as the infectious spleen and kidney necrosis virus contain both TNs in their DNA as TNs of minimum and approximately equal concentrations (CTAGmin~ TCGAmin). The same characteristic is observed in the virus Ranid 1 of the family Alloherpesviridae. DNAs of most of the alpha-herpeviruses of the genus Simplexvirus contain these tetramers at minimum concentrations as CTAGmin < TCGAmin.
The concentration of TCGA is not minimum in DNAs of roseoloviruses. At the same time, most of the herpesvirus DNAs (26 out of 35) — except for gamma-herpesviruses — have CTAG at minimum concentrations. Section 2 describes distinctive characteristics of nTAn and nCGn FPs in DNAs of herpesviruses. The minimum concentration of CTAG is observed in all the studied Nucleocytoviricota — except for poxviruses, among which only 3 chordopoxviruses (out of 19 analyzed viruses) have CTAGmin. As was said, poxviruses have DNA with dominant type A+T and a high ratio (> 2) ratio of [T+A]:[G+C].
Before we move to the next section, we would like to point out the key aspects:
1) The DNA of herpesviruses is different from DNAs of other viruses discussed here by the ratio of [G+C] > [A+T].
2) Among the studied viral DNAs, the TCGA tetramer is present at minimum concentrations almost exclusively in DNAs of herpesviruses – with more than two-fold dominance of G+C over A+T. These are, first of all, herpesviruses of the genus Simplexvirus of the subfamily Alpha and, partially, of the genus Lymphocriptovirus of the subfamily Gamma. DNAs of many herpesviruses have the ACGT tetramer represented at minimum concentrations; however, it is not unique, as its minimum concentrations are found in DNAs of other classification groups of viruses.
2. FREQUENCY PROFILE OF TETRA-NUCLEOTIDES IN VIRAL DNAS
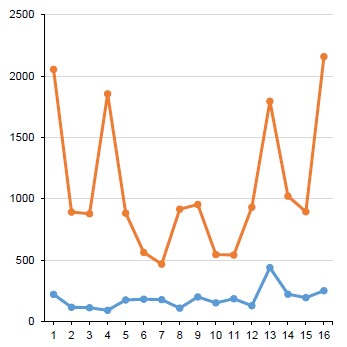
The symmetry of their FPs is a distinctive feature of quantitative distribution of nCGn genomic tetramers of some viruses of the phylum Peploviricota. Fig. 1 shows it for the herpes simplex virus type 1 whose genome is organized by type D [18]. The nTAn FP is asymmetrical due to the minimum concentration of CTAG (CTAGmin). The ratio CTAG:GTAC and TCGA:ACGT is more precise than symmetry, though also representing it. For DNA of the herpes simplex virus type 1, it is CTAGmin< GTAC (21%) and TCGAmin< ACGT (86%).
Fig. 1. FPs of nTAn (blue) and nCGn (red) tetra-nucleotides in Human Simplexvirus 1 DNA. a — absolute values; the percentage ratio of symmetric pairs of two complete tetramers — TCGAmin:ACGT and CTAGUNP:GTAC (shown in green and bold); b, c — a graphic representation of absolute values; c — nTAn FP; vertical scale up.
Once the central dimer of the CTAG tetramer is replaced by the reverse one — CATG, the symmetry of the respective FP is restored. The asymmetry of nCGn FPs is much less pronounced, though the TCGA<ACGT ratio is quite typical of FPs of many simplex viruses. Any substitutions of the CG central dimer result in severely distorted and broken FP symmetry.
Fig. 2 shows FPs of nTAn and nCGn tetra-nucleotides in the Human Cytomegalovirus (HHV5) DNA organized by type D, similarly to herpesviruses of the genus Simplexvirus. As can be noticed, the symmetries of both FPs are almost absent; however, CTAGmin:GTAC and TCGAmin:ACGT ratios are clearly presented (36 and 52%, respectively). It is obvious that the symmetry of nCGn FPs is associated with the GC-type DNA of the herpesvirus of the genus Simplexvirus.
Fig. 2. Frequency profiles of nTAn (blue) and nCGn (red) tetra-nucleotides in the Human Cytomegalovirus (HHV5) DNA.a — absolute values; b — their graphic representation. The percentage ratio of symmetrical pairs of two complete tetramers, ACGT:TCGA and GTAC:CTAG (shown in green and in bold).
The numeric GC:AT ratio and its association with symmetries of herpesvirus DNA FPs are illustrated by Table. Pronounced symmetries of nCGn FPs result from ratio GC:AT>2, which is typical of DNAs of herpesviruses of the genus Simplexvirus. Ratios of pairs TCGA2 (poxviruses).
Fig. 3 shows CTAG and TCGA FPs in DNA of Ranid herpesvirus 1, (genus Batrachovirus), in which both TNs have lowest FPs, while the genome is organized by V. Roizman’s type. In DNA of this virus, FPs of both TNs do not demonstrate symmetries; however, they quite clearly show the so-called incomplete tetramers — CTA/TAG and CCG/CGA trimers — by the decreasing density. To an extent, it can be observed in some other viral DNAs. Normally, the density of such trinucleotides does not reach levels of complete CTAG or TCGA.
Fig.3. Frequency profiles of nATn and nGCn tetra-nucleotides in DNA of Ranid herpesvirus 1. a — absolute values; 3’-trimers TAG and CGA are highlighted in green and bold; 5’-trimers CTA and CCG are underlined; below — the percentage ratio of complete tetramers: ACGT:TCGA and GTAC:CTAG; b, c — a graphic representation of absolute values; c — nTAn, vertical scale up
Summarizing the above section, we would like to point out two pronounced, though previously unaddressed, formal characteristics of CTAG and TCGA.
FPs of nCGn in DNAs [G+C]:[A+T]>2 demonstrate a certain symmetry, while the symmetry of nATn FPs in such DNAs is often broken (CTAG<GTAC). This symmetry does not follow Chargaff’s second parity rule, which, at least, is free of such limitations, and it does not stem from this rule, as it may look on the surface. It is also different from symmetries of DNA sequences, which were described in the following works [19–21].
Not only CTAG, but also its trinucleotide overlapping regions, nTAG and CTAn, have, as a rule, a more or less pronounced tendency toward decreased density in FPs of the respective tetramers (Fig. 2). The tendency toward lower density is also demonstrated by nCGA and TCGn trimers. If nTAG or CTAn trimers overlapping the 5’ or 3’-regions of CTAGmin are seen as those of the minimum concentration, the number of 75 CTAGmin out of 128, which we referred to when discussing DNAs of the representatives of phylum Uroviricota, will increase to 93. Such trinucleotides also demonstrate a tendency toward decreased frequency, which generally does not reach minimum values of the complete CTAGmin. Therefore, we think that the above-mentioned hypothesis of the thermodynamic model, which applies to the complete CTAG TN, needs some clarification.
Characteristics of the 1st (A) and 3rd (G) lines of virus (mainly herpesvirus) genomes with ratio GC : AT > 1
Note. Tetra-nucleotides (nCGn) of the minimum concentration are shown as ACGT or TCGA. The FP symmetry is shown in grey color (see the explanation in the text).
These four trinucleotides (CTA/TAG partially overlapping CTAG and TCG/CGA, partially overlapping TCGA) have another surprising feature, which, at first glance, is not associated with their known functions; in fact, it falls into the scope of the next section of the article, and is mentioned here as a transition to it: these four trimers, which are seen as overlapping codons, exhaust the excessiveness of the universal genetic code (shown in bold; Roman numerals are used to denote the group of their degeneracy): CTAIV = TTRII(L) and TAGII = TGAIII(stop); TCGIV = AGYII(S) and CGAIV = AGRII(R). The group of their degeneracy is always higher than the group of degeneracy of alternative codons of the same amino acid.
Alternative codons constitute the symmetrical central line A-T-T-T-A (the first letters) or SII-stop-LIIstop-RII (coding products) of the so-called code matrix [17]. In the genetic code, there is not any pair of complete self-complementary TNs like TCGA and CTAG, which would be overlapped by trinucleotides with similar properties. This distinctive characteristic, which can be of a casual nature, prompts to take a closer look at he structure of the universal genetic code in the above context and share the observations that are not always explainable, but deserve attention.
3. UNIVERSAL GENETIC CODE AND GENOMIC SYMMETRIES OF TCGA AND CTAG TETRA-NUCLEOTIDES
The genetic code owes the robustness of its structure mainly to the symmetry of its elements. Rumer’s table is one of the most illustrative examples of such symmetry (the first in the history) [22][23]; it was later converted by V. Scherbak into a calligram, with the symmetry between the first letters of coding triplets and the coding products arranged by their molecular weight [16]. This symmetrical relationship does not still have any clear explanation. In our slightly modified table (calligram A; Fig. 4, a), we placed an emphasis on the evolutionary stages of the code — as opposed to the established degeneracy groups in the original calligram. It shows the dominance of G+C in the first octet, which, apparently, reproduces the dominance of more thermally stable pairs G≡C in the “pre-code” set of polynucleotides, and A+T in the second octet, which is also responsible for gene reading and other features, thus being more complex and, most likely, evolutionary younger. In our version, octet 2 of the calligram (Fig. 4, b) is “packed tight” as the amino acid sequence is based on the total weight of products encoded by triplets with the third pyrimidine Y or purine R. This octet is referred to as octet A (Fig. 4, a) by the prevailing total content of nucleotides A in the 1st and 2nd coding lines. For the same reason, octet 1 of the calligram is referred as octet C. The number of all four nucleotides in the first lines of each octet is identical, thus emphasizing the inter-octet symmetry.
Fig. 4. Calligram A of the universal genetic code (a) and octet 2 of the calligram of the universal genetic code ([16]; b).The third nucleotide of the codon is represented by purine (R) or pyrimidine (Y). The start codon ATG and the stop codon TAR are highlighted vertically in grey. The first letters of central tetra-nucleotide codons of each octet are shown in bold. The tetra-nucleotide of the junction of octets A and C is also highlighted in grey. The successive increase in the molecular weight of the coding products (AC, amino acids) is shown by the increasing background density from white to black and by arrows. Roman numerals are used to denote degeneracy groups of the code, dg. Three pairs of coding products, some of which can bear a charge (that is why their positions in the lines are not stable and fixed following the dominant rule — the symmetry of the first letters of codons), highlighted in light grey
Octet C in calligram A is organized by the successive change (increase) in the molecular weights of encoded amino acids; such organization results in the symmetry of the upper line nucleotides (the first letters of codons). The TCGA tetramer is the core of this symmetry (shown in bold on the grey background). We omit the values of molecular weights of coding products; they can be found in the following works [15][16]. Octets C and 1 of both calligrams are completely identical.
Octet A in our calligram is also based on the successive change (though this time – toward a decrease) in molecular weights of encoded amino acids, thus having the symmetry of the upper line nucleotides (the first letters of codons). Amino acids comprising three pairs — R+S, E–D– and K+N (in Fig. 4 they are highlighted in light grey) have quite similar molecular weights, which may vary due to their ability to bear a charge (protonation). However, the dominant principle of the octet organization, namely, the symmetry of the first letters of coding triplets secures the central CTAG tetramer in the upper line of octet A, placing the glutamic acid in the fourth position of the tetramer. A certain role here may belong to the symmetry of charges of histidine (H+) and glutamine (D–) at a neutral pH, with third codon letters represented by pyrimidines. These two factors — successive changes in the molecular weight of encoded products and the symmetry of the first letters of codons — set the direction of the octet reading and the direction of gene reading — from triplet ATG (start codon) to triplets TGA and TAR (stop codons).
Both octets of the genetic code can represent its presumed evolution [7][26] — from occasional start/ stop codons of octet C to fixed (octet A) codons and from the dominance of G and C nucleotides in octet C to their alignment due to dominating A and T nucleotides in codons of octet A, thus making the octet more complex.
The approach we mentioned at the end of the previous section, namely, tetramers being overlapped by trimers, increases the information content of the calligram, showing even the odd-numbered groups of degeneracy. The linear four-nucleotide “junction” of the first lines of octets A and C, i.e. AT|GG, can be seen as being partially overlapped by ATG and TGG codons of the degeneracy group I (shown in grey in Fig. 4, a). The presence of this junction demonstrates the multidirectional organization of octets A and Cght, which produces their symmetry — decreasing or increasing of the nucleon mass of coding products with unidirectional central octet tetramers.
The analysis of nCGn and nTAn tetramer FPs in chains of the 1st, 2nd and 3rd lines of viral genomes reveals a certain similarity with symmetries of these FPs in the 1st, 2nd and 3rd lines of the genetic code. The first chain starts with nucleotide A, the second chain starts with T, and the third one starts with G, while genes are arranged one after another, without spacing, regardless of viral DNAs, overlapping and introns. The examples are given only for genomes of the viruses discussed in the previous section: HHV1 (Fig. 5), HHV5 (Fig. 6) and RaHV-1 (Fig. 7).
Fig. 5. FPs of nCGn (a) and nTAn (b) tetramers in chains of the 1st, 2nd and 3rd nucleotides of the Herpes simplex virus 1 genome.Here and in Fig. 6, 7: on the left — absolute values, on the right — their graphic representation in chains of the 1st, 2nd and 3rd nucleotides to demonstrate proportions of the profile (but not its scale, which can be estimated with the help of absolute values presented in the numeric section of the figure).
Fig. 6. FPs of nCGn (a) and nTAn (b) tetramers in chains of the 1st, 2nd and 3rd nucleotides of Human cytomegalovirus (HHV5) genome.
Fig. 7. FPs of nCGn (a) and nTAn (b) tetramers in chains of the 1st, 2nd and 3rd nucleotides of Ranid herpesvirus 1 genome.
Figure 5 (a) demonstrates the result of the analysis – the symmetry of the chain of the first nucleotides of the HHV1 genome, in which the TCGA tetramer has the minimum concentration, giving rein to the ACGT tetramer. The symmetry of the second nucleotides is absent in the same way as it is absent in the 2nd line of the code calligram. Both facts are consistent with the functions of the 1st and 2nd nucleotides of the codon, and their nature (the presence and absence of a symmetry) correlates with the organization of the universal genetic code. The well-defined symmetry of the chain of the 3rd nucleotides of the genome, which may look as redundant, as the nucleotides are selected spontaneously, prompts the idea of compensation for the FP symmetry of the first nucleotides of the genome and the nCGn FP in the actual HHV1 DNA (Fig. 1). In addition, the similar symmetry could be typical of nucleotide polymers existing before the genetic code or selected for its evolution. The analysis of the complete herpesvirus DNA divided into three chains similarly to the genome restores the statistical nature of the nCGn FP, i.e. the similar symmetry of tetramers for all 3 chains without reference to genes.
Figure 5 (b) shows the heavily distorted symmetry of the nTAn FP in the chain of the first letters of codons — apparently, due to small numbers of nTAn tetramers. In fact, the same could be applied to TCGA, but its functional dimer (CG) has much more frequent occurrence even in the irregular chain compared to the functional CTAG tetramer and it can keep the illusion of the function much better with the tetramer than with the CnnTnnAnnG decamer. It should be noted that the TnnCnnGnnA decamer also has the minimum concentration. The shortage of CTAG in the chain of the first letters of coding triplets discontinues, though the concentration levels of CTAG are still slightly lower than those of the symmetric GTAC. Similar to nCGn (Fig. 5, a), the nTAn FPs of the second letters do not demonstrate symmetry, while the concentrations of the tetramers of the 3rd chain are so low that they can be ignored; nevertheless, they follow the order of the values of the 1st letters and may participate in the overall symmetry of nTAn FPs in the actual HHV1 DNA.
Figure 6 shows that FPs of the chain of the 1st, 2nd and 3rd letters of the nCGn tetramer (and nATn to a lesser extent) of the beta-herpesvirus HHV5 genome “restore” the symmetry absent in the actual DNA of this virus, while losing CTAG and TCGA as minimum concentrations. Similar to the HHV1 genome, the FP of the second letters of both tetramers lacks symmetry in the HHV5 genome.
Figure 7 presents FPs of the discussed tetramers in chains of the 1st, 2nd and 3rd letters of the Ranid herpesvirus 1 alloherpesvirus genome. Here, we can also see the “restoration” of the FP symmetry in the chains of the 1st and 3rd letters of the nCGn tetramer, including the nATn genome, though to a lesser extent — the symmetry that was absent in the actual viral DNA. The concentrations of CTAG and TCGA tetramers remain to be low, though their levels become much lower.
We summarized the obtained results in the table showing the data for nCGn FPs in DNAs of viruses with ratios of [G+C]:[A+T]>1.0. These viruses are primarily represented by herpesviruses. Two characteristics were addressed: the genus of Simplex herpesviruses with TCGAmin typical of their DNAs and the symmetry of the respective profile. To a larger extent, these characteristics are observed in simplex genomes or, to be more exact, the chains of their 1st (and 3rd) codon nucleotides.
The question arises about DNAs with the AT type and the similar high ratio of [A+T]:[G+C]. Among the studied viruses, such ratio is more frequently observed in poxvirus DNAs and genomes. The similarity with herpesviruses has been found only in symmetries of FPs of the analyzed tetramers and only when the order of the fringe bases of the quadruple changes from CTAG to TCGA.
Summarizing the aforesaid, we would like to point out several formal characteristics of TCGA and CTAG tetramers with reference to the genetic code structure.
TCGA (octet C) and CTAG (octet A) are central tetramers of the first lines of octets in calligram A.
For the GC genome type, FPs of nCGn and nTAn tetramers demonstrate the bilateral symmetry of the 1st and 3rd lines of nucleotides in genomes of a number of viruses and the absence of such symmetry in lines of the 2nd nucleotides. These characteristics of lines are also typical of the genetic code. The FPs of the 3rd lines (G) of genomes demonstrate the symmetry at lower limitations [G+C]:[A+T]>1. The FPs of the 1st, 2nd and 3rd chains of complete (not limited by the genome) DNAs whose genes are not identified offset the observed difference between the chains of the genome.
The FP of nCGn herpesvirus DNAs shows the TCGA tetramer (remember that it is not the TN in the final version of the code) as the least represented in lines A and G in most of the studied cases, while the FP of nTAn does not show herpesviruses as the group with unique CTAGmin concentrations compared to other groups of viruses.
With sizes of all three lines of the viral genome (GC type of DNA) being naturally equal, the total number of nCGn tetramers of the 1st and 2nd lines is approximately equal to the number of such tetramers in the 3rd line.
Thus, both groups of functional TNs — TCGA and CTAG described in sections 2 and 3 share the common characteristic — the symmetry found both in complete viral DNAs and in individual codon lines of genomes of these viruses. In the first case, it refers to DNAs of “current” viruses; in the second case, it refers to their genomes and to the genetic code. Both groups of symmetries, including their arrangement, bring up a question about the origin of viruses or, at least, about the origin of some of them.
DISCUSSION
Life on Earth started from glycosylation and phosphorylation of purines and pyrimidines with the further selection of uniform optical isomers and their non-template polymerization. None of these processes — in existing natural conditions on our planet — can take place without enzymes, though during early stages of abiogenesis, enzymes could have been replaced by different clays [24]. The events and factors that prepared (on the planet or even outside the Earth system), launched and scaled up the abiogenetic process more than 4 billion years ago remain the subject of much speculation; the question whether everything could have happened by accident also remains unanswered [25][26]. The further evolution could depend on clusters of microscopic compartments (also with participation of the above-mentioned clays), inside which growing heteropolymers competed for limited resources. The “losers” were destroyed and were used by the “winners” or were driven out the compartment through its semipermeable membrane. If they survived the aggressive external environment and were able to penetrate into the closest compartment or get into it after the fusion, they continued fighting with new competitors and that time their fight could be successful. In terms of compartments, the behavior of these competitors was very similar to the behavior of current viruses, though the compartment was highly different from the modern cell. The “winner’s” advantage depended on the growth rate within the limits of permissible dimensions and on the evolving template replication catalyzed by ribozymes — products of the RNA world [27][28], proto-metallopolyproteins [29] or random factors.
During that stage, the described events developed along two clearly defined lines: intense competition among the participants for the growth resources and the evolution of the system required by this competition. The stability of polymers could be supported by their structure with a double chain during the inter-replication period [30]; the total length of the chain was preserved, while single-chain sections more sensitive to damage were reduced and there were multiple repeats contributing to the symmetry of the chain. This system may have emerged repeatedly for short periods in different areas on the planet, but eventually it approached the fundamental evolutionary leap when the translation machinery and genetic code were created to stabilize the cooperation of nucleotide and amino acid heteropolymers and significantly reduce the randomness of further processes at the molecular level.
Nucleotide polymers capable of growth and replication stored the information defining the amino acid sequences that were able to catalyze synthesis and replication processes much more efficiently than random factors during the previous stages.
The genetic code stabilized the life chemistry and significantly accelerated its evolution leading to organization of the first cells and dividing positions of nucleic acids into intracellular and extracellular, thus securing the first two central biological elements capable of efficient interaction (competition or cooperation) — the cell and the virus, which gained a possibility to grow in size. Viruses, most likely, continued to evolve further on and through other ways [31–34].
Some scientists believe that the genetic code evolved in stages [35–38]. We assume that initially, the code continued to have characteristics of “pre-code” heteropolymers, including some excessive concentrations of G and C, as well as some symmetry elements (due to repeats) increasing its robustness. The basis of the code symmetry was formed not only by complementarity, but also by another parameters combining codons and coding products — the molecular weight (the size) of participants. The CpG dimer, which due to its abundance, most likely, became the initial structural element of the code, is characterized by complementarity of C≡G and the ratio of C˂G (Y˂R) for molecular weights of monomers. This dinucleotide might be performing some unique functions in synthesis of biopolymers, thus standing out among others and, therefore, being selected as the initial element. Some scientists assume that the first codons were doublet [35]. Later, the Y˂R ratio was preserved and the set of first nucleotides of the code was extended to the full four-letter size — TCGA.
At a later stage, the Y˂R ratio formed the basis for the assembly of another tetramer — CTAG, which (this time acting as TN) also had a unique biological function. This tetramer specified the unidirectionality of ratios between nucleotides from the pyrimidine-purine level to the level of nucleotides (C˂T˂A˂G).
The evolution of the size of codons initially resulted in mutual overlapping, which later was replaced by the triplet structure of the code with different functions of the 1st, 2nd and 3rd letters of the codon. The first letters were responsible for the code stability provided by the symmetry, which was based on the successively changing molecular weights of coding products. Amino acids, which share the same biosynthetic pathway, normally share the first position in codons [25]. The second letters of coding triplets are responsible for functions of amino acids depending on their polarity; codons of amino acids with similar physical and chemical properties are usually also characterized by similarity, thus helping alleviate consequences of point mutations and impaired translation. The third letters of codons separate coding doublets with purines or pyrimidines (octet A) or with their random selection (octet C) [23]. Based on the above organization, both tetramers belong to different groups of degeneracy, which continued to exist even outside their boundaries.
The product of the code evolution was the predecessor of octet C (the dominance of C and G) and later (or concurrently), when the code acquired additional features – direction of the gene reading and codon differentiation by the third letters, purine or pyrimidine – emergence of octet A (compensatory dominance of A and T; Fig. 4).
Existing “live” single-stranded nucleic acids (separate chains of genomes) also demonstrate a certain symmetry, including the symmetry of FPs of functional TNs. Viral DNAs are the best choice for studying this symmetry, as their genome, i.e. a set of genes encoding sequences, occupies the largest portion of the DNA (more than 80% in herpesviruses).
We demonstrated that in the genome of herpesviruses with high concentrations of GC, the nCGn FPs in the chains of the 1st, 2nd and 3rd nucleotides are characterized by the symmetries similar to those observed in the chains of the 1st and 3rd nucleotides of the genetic code. On the other hand, the nCGn FPs in the chain of two-codon nucleotides do not have such symmetry, though they share the same characteristics with the other two chains in addition to the common unseparated DNA strand: Type GC and ratio [G+C]:[A+T] > 2. The differences in nCGn FPs in the chains of the 1st, 2nd and 3rd letters of the genome correspond to the functions of codon nucleotides and the formal structure of the genetic code. The total number of nCGn tetramers in the 3rd chain of the viral genome with high concentrations of GC is approximately equal to the total number of nCGn tetramers in the 1st and 2nd chains (codons of octet C mean the choice out of 2 with the first two nucleotides and the choice out of 4 with the third nucleotide).
The aforesaid illustrates the functional character of calligrams of the genetic code, which are more informative than the standard table and its versions available in most students’ books.
We assume that the symmetry of FP of nCGn nucleotides in the third chain — similarly to the general symmetry of nCGn FPs — can be an atavistic feature of the pre-code pool of polynucleotides. On the other hand, the properties of the third chain can be required as a “reserve” to provide the symmetry of the first chain. Undoubtedly, the described symmetries could be formed by any, even by randomly generated DNA polymers of sufficient length. However, in this case, when they were divided into three chains following the above principle, the second chain would not be identified by such symmetries.
The symmetry of nCGn FPs preserved at least in one of the three chains of the viral genome after it is split up means that under specific conditions, the limit set for the size of the genome can be increased approximately 2–3-fold compared to the limit we set at the beginning of our study (100 kbp).
Most certainly, the emphasis placed on herpesviruses in our article (and on adenoviruses too) does not lead to the assumption that life on Earth started from the above viruses. These viruses, their components (and their hosts) are too complex both structurally and functionally [18], and their DNA is too large, implying that they have gone through long evolution, which involved their type (GC) and high GC/AT ratio [39]. This evolution involved not only DNA, but also coding products — proteins, more stable components of life [40][41]. It is individual proteins that demonstrate evolutionary relatedness between herpesviruses and tailed phages when compared in different viruses [42], while the DNA structure shows quite a few evolutionary discrepancies in the discussed parameters. The role of terminal repeats in DNAs is not as apparent for its contribution to symmetries, though they also can be of atavistic, relict nature.
Symmetries of the genetic code have been discussed before and they keep attracting attention of scientists studying them from different perspectives [43][44]. Here, we take a closer look at one of the aspects of these symmetries.
By publishing these data, we wanted to point out the characteristics and similarities of two biological objects, which are seemingly unrelated, though they have common and significant markers — TCGA and CTAG tetramers, including such quality if their FPs as symmetries. The first object is the viral (in our case) DNA; the other object is the universal genetic code. The presented data suggest an evolutionary relationship between these objects, which is based on poorly studied biological functions of these tetramers. Although these functions are seemingly different in the DNA biosynthesis and in the process of code evolution, such differences may have stemmed from the conditions of their emergence at different stages of the biological evolution.
1. URL: https://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=10239&sort=taxonomy
About the authors
Felix P. Filatov
I. Mechnikov Research Institute of Vaccines and Sera; National Research Center for Epidemiology and Microbiology named after Honorary Academician N.F. Gamaleya
Author for correspondence.
Email: felix001@gmail.com
ORCID iD: 0000-0001-6182-2241
D. Sci. (Biol.), leading researcher, Laboratory of molecular biotechnology, Department of virology; leading researcher, Department of epidemiology
Russian Federation, Moscow; MoscowReferences
- Филатов Ф.П., Шаргунов А.В. Тетрануклеотидный профиль герпесвирусных ДНК. Журнал микробиологии, эпидемиологии и иммунобиологии. 2020; 97(3): 216-26. https://doi.org/10.36233/0372-9311-2020-97-3-3
- Tang L., Zhu S., Mastriani E., Fang X., Zhou Y.J., Li Y.G., et al. Conserved intergenic sequences revealed by CTAG-profiling in Salmonella: thermodynamic modeling for function prediction. Sci. Rep. 2017; 7: 43565. https://doi.org/10.1038/srep43565
- Lundberg P., Welander P., Han X., Cantin E. Herpes simplex virus type 1 DNA is immunostimulatory in vitro and in vivo. J. Virol. Oct. 2003; 77(20): 11158-69. https://doi.org/10.1128/JVI.77.20.11158-11169.2003
- Sharawy M., Louyakis A., Gogarten J.P., May E.R. CTAG vs. GATC: structural basis for representational differences in reverse palindromic DNA tetranucleotide sequences. Biophys. J. 2021; 120(3): 222a.
- Albrecht-Buehler G. Asymptotically increasing compliance of genomes with Chargaff's second parity rules through inversions and inverted transpositions. Proc. Natl Acad. Sci. USA. 2006; 103(47): 17828-33. https://doi.org/10.1073/pnas.0605553103
- Albrecht-Buehler G. The three classes of triplet profiles of natural genomes. Genomics. 2007; 89(5): 596-601. https://doi.org/10.1016/j.ygeno.2006.12.009
- Zhang S.H., Wang L. A novel common triplet profile for GCrich prokaryotic genomes. Genomics. 2011; 97(5): 330-1. https://doi.org/10.1016/j.ygeno.2011.02.005
- Stevens M., Cheng J., Li D., Xi M., Hong C., Maire C., et al. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 2013; 23(9): 1541-53. https://doi.org/10.1101/gr.152231.112
- Krieg A.M, Yi A.K., Matson S., Waldschmidt T.J., Bishop G.A., Teasdale R., et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995; 374(6522): 546-9. https://doi.org/10.1038/374546a0
- Fatemi M., Pao M.M., Jeong S., Gal-Yam E.N., Egger G., Weisenberger D.J., et al. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic. Acids Res. 2005; 33(20): e176. https://doi.org/10.1093/nar/gni180
- Woellmer A., Hammerschmidt W. Epstein-Barr virus and host cell methylation: regulation of latency, replication and virus reactivation. Curr. Opin. Virol. 2013; 3(3): 260-5. https://doi.org/10.1016/j.coviro.2013.03.005
- Burge C., Campbell A.M., Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. PNAS. 1992; 89(4) 1358-62. https://doi.org/10.1073/pnas.89.4.1358
- Duret L., Galtier N. The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is due to a mathematical artifact. Mol. Biol. Evol. 2000; 17(11): 1620-5. https://doi.org/10.1093/oxfordjournals.molbev.a02621.
- Gori F., Mavroeidis D., Jetten M.S.M., Marchiori E. The importance of Chargaff’s second parity rule for genomic signatures in metagenomics. bioRxiv. Preprint. https://doi.org/10.1101/146001
- Rudner R., Karkas J.D., Chargaff E. Separation of B. subtilis DNA into complementary strands, 3 Direct Analysis. Proc. Natl Acad. Sci. USA. 1968; 60(3): 921-2. https://doi.org/10.1073/pnas.60.3.921
- Makukov M.A., Shcherbak V.I. The “Wow! signal” of the terrestrial genetic code. Icarus. 2013; 224(1): 228-42. https://doi.org/10.1016/j.icarus.2013.02.017
- Filatov F. A molecular mass gradient is the key parameter of the genetic code organization. In: Blaho J., Baines J., eds. From the Hallowed Halls of Herpesvirology: A Tribute to Bernard Roizman. World Scientific Publishing Co.; 2012: 155-68. https://doi.org/10.1142/9789814338998_0006
- Pellett P., Roizman B. Herpesviridae. In: Knipe D.M., Howley P.M., eds. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2013: 1802-2
- Prabhu V.V. Symmetry observations in long nucleotide sequences. Nucleic Acids Res. 1993; 21(12): 2797-800. https://doi.org/10.1093/nar/21.12.2797
- Forsdyke D.R. Symmetry observations in long nucleotide sequences: a commentary on the discovery note of Qi and Cuticchia. Bioinformatics. 2002; 18(1): 215-7. https://doi.org/10.1093/bioinformatics/18.1.215
- Baisnee P.F., Hampson S., Baldi P. Why are complementary strands symmetric? Bioinformatics. 2002; 18(8): 1021-33. https://doi.org/10.1093/bioinformatics/18.8.1021
- Румер Ю.Б. О систематизации кодонов в генетическом коде. Доклады Академии наук СССР. 1966; 167(6): 1393-4.
- Волькенштейн М.В., Румер Ю.Б. О систематике кодонов. Биофизика. 1967; 12(1): 10-3.
- Kim H.Y., Cheon J.H., Lee S.H., Min J.Y., Back S.Y., Song J.G., et al. Ternary nanocomposite carriers based on organic claylipid vesicles as an effective colon-targeted drug delivery system: preparation and in vitro/in vivo characterization. J. Nanobiotechnology. 2020; 18(1): 17. https://doi.org/10.1186/s12951-020-0579-7
- Koonin E.V., Novozhilov A.S. Origin and evolution of the genetic code: the universal enigma. IUBMB Life. 2009; 61(2): 99-111. https://doi.org/10.1002/iub.146
- Marlaire R., ed. Ames Research Center. NASA Ames Reproduces the Building Blocks of Life in Laboratory. Moffett Field, CA: NASA; 2015.
- Herbert K.M., Nag A. A tale of two RNAs during viral infection: how viruses antagonize mRNAs and small non-coding RNAs in the host cell. Viruses. 2016; 8(6): 154. https://doi.org/10.3390/v8060154
- Tjhung K.F., Shokhirev M.N., Horning D.P., Joyce G.F. An RNA polymerase ribozyme that synthesizes its own ancestor. Proc. Natl Acad. Sci. USA. 2020; 117(6) 2906-13. https://doi.org/10.1073/pnas.1914282117
- Kim J.D., Senn S., Harel A., Jelen B.I., Falkowski P.G. Discovering the electronic circuit diagram of life: structural relationships among transition metal binding sites in oxidoreductases. Philis. Trans. R Soc. Lond. B. Biol. Si. 2013; 368(1622): 20120257. https://doi.org/10.1098/rstb.2012.0257
- Yakovchuk P., Protozanova E., Frank-Kamenetskii M.D. Basestacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006; 34(2): 564-74. https://doi.org/10.1093/nar/gkj454
- Forterre P. The origin of viruses and their possible roles in major evolutionary transitionsa. Review. Virus Res. 2006; 117: 5-16.
- Mughal F., Nasir A., Caetano-Anollés G. The origin and evolution of viruses inferred from fold family structure. Arch. Virol. 2020; 165(10): 2177-91. https://doi.org/10.1007/s00705-020-04724-1
- Brussow H., Kutter E. Genomics and evolution of tailed phages. In: Kutter E., Sulakvelidze A. eds. Bacteriophages: Biology and Applications. Boca Raton, London, New York, Washington: CRC press; 2005: 129-64.
- Abedon S.T. Phage evolution and ecology. Adv. Appl. Microbiol. 2009; 67: 1-45. https://doi.org/10.1016/s0065-2164(08)01001-0
- Altstein A.D. The progene hypothesis: the nucleoprotein world and how life began. Biol. Direct. 2015; 10: 67. https://doi.org/10.1186/s13062-015-0096-z
- Di Giulio M. The origin of the genetic code: theories and their relationships, a review. Biosystems. 2005; 80(2): 175-84. https://doi.org/10.1016/j.biosystems.2004.11.005
- Gilis D., Massar S., Cerf N.J., Rooman M. Optimality of the genetic code with respect to protein stability and amino-acid frequencies. Genome Biol. 2001; 2(11): RESEARCH0049. https://doi.org/10.1186/gb-2001-2-11-research0049
- Wetzel R. Evolution of the aminoacyl-tRNA synthetases and the origin of the genetic code. J. Mol. Evol. 1995; 40(5): 545-50. https://doi.org/10.1007/bf00166624
- McGeoch J., Rixon F.J., Davison A.J. Topics in herpesvirus genomics and evolution. Virus Res. 2006; 117(1): 90-104. https://doi.org/10.1016/j.virusres.2006.01.002
- Wang N., Baldi P.F., Gaut B.S. Phylogenetic analysis, genome evolution and the rate of gene gain in the Herpesviridae. Mol. Phylogenet. Evol. 2007; 43(3): 1066-75. https://doi.org/10.1016/j.ympev.2006.11.019
- Wertheim J.O., Smith M.D., Smith D.M., Scheffler K., Kosakovsky Pond S.L. Evolutionary origins of human herpes simplex viruses 1 and 2. Mol. Biol. Evol. 2014; 31(9): 2356-64. https://doi.org/10.1093/molbev/msu185
- Baker M.L., Jiang W., Rixon F.J., Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005; 79(23): 14967-70. https://doi.org/10.1128/JVI.79.23.14967-14970.2005
- Гупал А.М., Гупал Н.А., Островский А.В. Симметрия и свойства записи генетической информации в ДНК. Проблемы управления и информатики. 2011; 5(3): 120-7.
- Сергиенко И.В., Гупал А.М., Вагис А.А. Симметричный код и генетические мутации. Кибернетика и системный анализ. 2016; (2): 73-80.
Supplementary files