Том 98, № 6 (2021)
- Год: 2021
- Выпуск опубликован: 10.01.2022
- Статей: 11
- URL: https://microbiol.crie.ru/jour/issue/view/47
Весь выпуск
ОРИГИНАЛЬНЫЕ ИССЛЕДОВАНИЯ
Методика оценки активности полифункционального белка трансферринового ряда при экспериментальном моделировании кинетики развития Staphylococcus aureus
Аннотация
Введение. Лактоферрин (ЛФ) представляет собой катионный мономерный гликопротеин, вырабатываемый ацинарными клетками и железами. ЛФ присутствует в разных местах слизистой оболочки в различной концентрации. В связи с разработкой различных вариантов гигиенических и лекарственных средств для лечения воспалительных заболеваний полости рта на основе ЛФ возникла необходимость объективной оценки его антибактериальных и антибиоплёночных свойств с последующим анализом сохранения активности при различных вариантах выделения данного белка из субстрата и хранения.
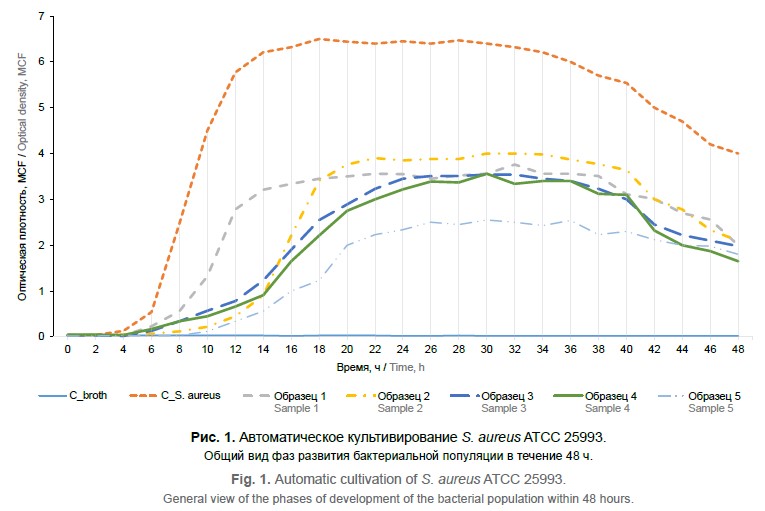
Цель исследования - повышение эффективности оценки антибактериальной активности ЛФ и продолжительности её сохранения в различных биологических субстратах, содержащих действующее вещество, и отдельных опытных партиях изготовленного препарата с помощью автоматического культивирования. Материалы и методы. В рамках эксперимента использовалась техника микробиологической диагностики с использованием системы автоматического культивирования микробных популяций. Заранее подготовленную бактериальную взвесь инокулировали в питательный бульон и добавляли исследуемые образцы ЛФ с последующим культивированием и анализом возможного антибактериального воздействия трансферринового белка. Для определения чувствительности выделенных штаммов применяли собственную модификацию метода серийных разведений, разработанную на кафедре микробиологии, вирусологии, иммунологии МГМСУ им. А.И. Евдокимова. В работе была использована инфраструктура Уникальной научной установки «Трансгенбанк». Результаты интерпретировали по изменению оптической плотности на длине волны λ = 850 нм. Изучение динамики роста микроорганизмов проводили в нескольких параллелях. Рост бактерий приводил к изменению параметров оптической плотности, на основании которых были построены кривые роста.
Результаты и обсуждение. По результатам экспериментального исследования кривых роста бактериальных популяций отмечены статистически достоверные различия количества жизнеспособных клеток в разные фазы кривых роста при использовании различных образцов ЛФ. Установлена более высокая активность образцов человеческого рекомбинантного ЛФ. При анализе динамики роста выявлены различия в наступлении максимума размножения и его ингибирования при воздействии различных отягощающих факторов в процессе культивирования. Бактериостатическое действие ЛФ реализуется посредством связывания ионов железа, лишая бактерии этого микроэлемента, вызывает ингибирование их развития. Наряду с этим ЛФ проявляет активность против некоторых факторов вирулентности микроорганизмов, расщепляя их по типу сериновых протеаз, и таким образом препятствует их проникновению в клетки человека.
Заключение. Использованная методика автоматического культивирования микроорганизмов в биореакторе позволяет получить воспроизводимые результаты, доступна для широкого использования и может быть рекомендована для получения объективных, сравнимых между собой, достоверных сведений о противомикробных свойствах различных образцов бактерицидного белка ЛФ, выпускаемых отечественной фарминдустрией. Исследуемый субстрат, содержащий рекомбинантный человеческий ЛФ российского производства, характеризуется высокой антибактериальной активностью, сохраняющейся, как минимум, в течение 3 лет.
 617-627
617-627


Применение секвенирования следующего поколения для исследования двойной ВИЧ-инфекции
Аннотация
Введение. Целью исследования был сравнительный анализ эффективности выявления и подтверждения двойной ВИЧ-инфекции с применением классического популяционного секвенирования (ПС) и секвенирования следующего поколения (NGS) для фрагмента гена pol ВИЧ-1, кодирующего протеазу и часть обратной транскриптазы (позиции 2253–3368).
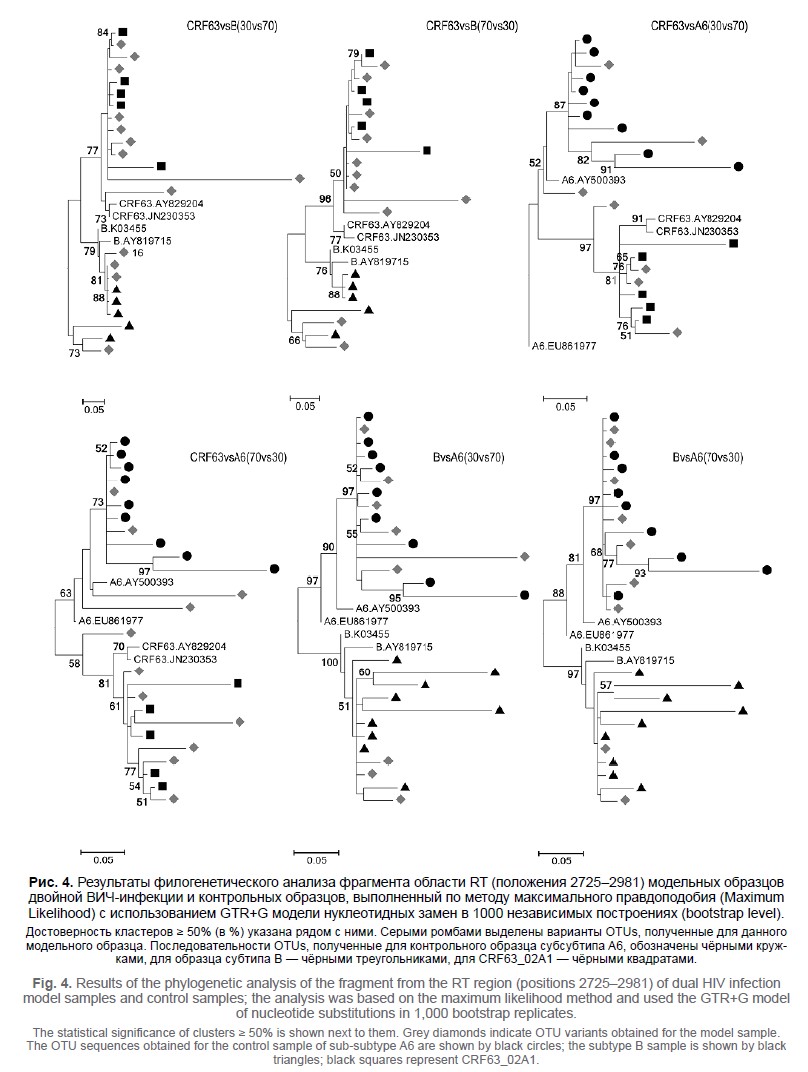
Материалы и методы. Исследованы модельные образцы межсубтиповой двойной ВИЧ-инфекции, содержащие вирусы ВИЧ-1 субтипа B, субсубтипа A6 и рекомбинантной формы CRF63_02A1. Вирусы смешивали попарно в соотношении от 10 до 90% для создания 3 групп модельных образцов: CRF63vsB, CRF63vsA6 и A6vsB. Нуклеотидные последовательности, полученные технологиями ПС и NGS, с порогами чувствительности к минорным вариантам вируса 5, 10, 15 и 20% (NGS5–NGS20 соответственно) использовали для определения величин индекса вырожденности (DB) и индекса синонимичности (SM). Фрагмент исследуемого региона (позиции 2725–2981) служил для анализа операционных таксономических единиц.
Результаты. Применение NGS5 оказалось наиболее эффективным для выявления двойной ВИЧ-инфекции в модельных образцах. Было обнаружено статистически достоверное (p < 0,01) увеличение DB- и SM-индексов для NGS5 по сравнению с ПС. Это позволило с помощью NGS5 выявить двойную ВИЧ-инфекцию в 25 модельных образцах из 27, в то время как ПС позволяло выявлять её лишь в 15 образцах. Анализ операционных таксономических единиц подтвердил двойную ВИЧ-инфекцию во всех группах модельных образцов.
Обсуждение. Эффективность выявления и подтверждения двойной ВИЧ-инфекции зависит как от доли каждого вируса в образце, так и от генетических особенностей данных вирусов. Заключение. Внедрение в рутинную практику генетического анализа технологии NGS позволит не только более эффективно выявлять генетические особенности инфекционных агентов, но и проводить более глубокий анализ эпидемиологической ситуации.
 627-638
627-638


Фенотипический профиль актуальных полирезистентных сиквенс-типов (ST 1167, ST 944, ST 208) Аcinetobacter baumannii
Аннотация
Введение. Ежегодно в мире регистрируют порядка 1 млн случаев инфекций, вызванных Acinetobacter spp., что составляет 1,8% всех случаев внутрибольничных инфекций. Клинически значимым представителем рода Acinetobacter является Acinetobacter baumannii, о чем свидетельствуют результаты многолетних исследований, проведённых в нашей стране и за рубежом. Внутривидовое типирование микроорганизмов является неотъемлемой частью работы клинического микробиолога при расшифровке вспышек гнойно-септических инфекций в рамках эпидемиологического надзора за инфекциями, связанными с оказанием медицинской помощи. Большинство практикующих микробиологических лабораторий не имеют возможности использовать генотипические методы типирования в силу высоких финансовых затрат.
Цель - разработка диагностической панели для внутривидовой идентификации A. baumannii сиквенс-типов ST 1167, ST 944, ST 208 на основе фенотипических свойств.
Материалы и методы. Изучены внутривидовые признаки 74 штаммов A. baumannii из 4 многопрофильных медицинских организаций крупного промышленного центра с использованием генетических (мультилокусное сиквенс-типирование) и комплекса фенотипических методов (биохимические тесты, биоплёнкообразующая способность, зоны задержки роста вокруг дисков с антибактериальными препаратами, чувствительность к анилиновым красителям, дезинфицирующим средствам и ацинетобактерному бактериофагу). Результаты. Определены фенотипические характеристики 3 ведущих сиквенс-типов A. baumannii (ST 1167, 944, 208). Обсуждение. Сформирован эффективный и экономичный набор дифференцирующих тестов, позволяющих выявить внутривидовые особенности полирезистентных штаммов A. baumannii актуальных сиквенс-типов.
Заключение. Диагностическая панель позволит проводить внутривидовую дифференциацию трёх широко представленных сиквенс-типов A. baumannii в рамках микробиологического мониторинга инфекций, связанных с оказанием медицинской помощи.
 639-647
639-647


Формирование специфического иммунитета у лабораторных животных после одновременной вакцинации против сезонного гриппа и COVID-19
Аннотация
Введение. Клиническая дифференциальная диагностика COVID-19 может быть затруднительна в случае совпадения с сезоном гриппа, что, в свою очередь, может приводить к несвоевременности принятия необходимых мер для борьбы с пандемией SARS-CoV-2. Существует также проблема сопутствующего SARSCoV-2 инфицирования вирусом гриппа (ВГ), что значительно утяжеляет течение COVID-19. Целью настоящей работы было изучение взаимного влияния одновременной иммунизации отечественными вакцинами для профилактики гриппа и COVID-19 на формирование специфического иммунитета лабораторных животных.
Материалы и методы. В исследовании использовали мышей линии BALB/c. Иммунизацию животных проводили внутримышечно вакциной для профилактики COVID-19 (КовиВак) и вакциной для профилактики гриппа (Флю-М). Сыворотки иммунизированных животных исследовали индивидуально. Реакцию торможения гемагглютинации проводили с тремя штаммами ВГ. Антитела (АТ) к SARS-CoV-2 определяли при помощи иммуноферментного анализа. Для выявления вируснейтрализующих АТ к SARS-CoV-2 и к ВГ проводили реакцию нейтрализации.
Результаты. Обнаружены достаточно высокие титры специфических АТ в группах животных, привитых как одной, так и двумя вакцинами одновременно. В группах животных, привитых КовиВак и двумя вакцинами одновременно, как в иммуноферментном анализе, так и в реакции нейтрализации средние показатели специфических АТ к SARS-CoV-2 статистически не различались. В группе животных, привитых одновременно двумя вакцинами, обнаружены статистически более высокие титры АТ к ВГ после второй иммунизации относительно группы животных, привитых Флю-М.
Обсуждение. Продемонстрировано формирование поствакцинального иммунитета как к ВГ, так и к SARSCoV-2 после одновременной иммунизации двумя вакцинами. Обнаруженное усиление поствакцинально- го иммунного ответа к ВГ у лабораторных животных, привитых двумя вакцинами одновременно, требует дальнейшего изучения.
Заключение. Проведённые исследования позволяют предположить возможность одновременной вакцинации для профилактики гриппа и COVID-19.
 648-656
648-656


Анализ изменения состояния влагалищной микрофлоры у женщин репродуктивного возраста в условиях трёхсуточной «сухой» иммерсии без использования средств профилактики
Аннотация
Введение. В связи с возрастающим участием женщин в космических полётах встают вопросы о влиянии факторов полёта на состояние женского организма и, в частности, на стабильность, видовые и количественные изменения микрофлоры влагалища. Для изучения влияния отдельных факторов космического полёта на организм наиболее удобными являются модельные эксперименты, в частности, «сухая» иммерсия.
Целью данной работы является сравнительная оценка состояния микробиоты влагалища у 6 женщин-испытателей до и после 3-суточного пребывания в «сухой» иммерсии.
Материалы и методы. Всем испытателям выполнена микроскопия вагинального отделяемого, окрашенного по Граму, и культуральное исследование в соответствии с медицинской технологией «Интегральная оценка состояния микробиоты влагалища. Диагностика оппортунистических вагинитов». Видовую идентификацию микроорганизмов проводили методом MALDI-TOF-MS-анализа.
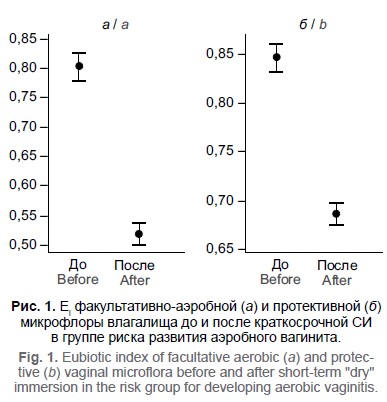
Для количественной оценки изменения состояния влагалищной микрофлоры и микрофлоры цервикального канала использовали эубиотический индекс, отражающий отношение числа положительных состояний микробиоты (до эксперимента — по отношению к норме, после эксперимента — по отношению к состоянию до эксперимента) к числу отрицательных.
Результаты. После 3-суточной «сухой» иммерсии у испытателей, которые до изоляции имели высокий титр аэробных микроорганизмов, количество аэробной микрофлоры существенно увеличилось, при этом количество протективных видов снижалось. У испытателей, которые до изоляции имели низкий титр аэробных микроорганизмов, количество аэробной микрофлоры уменьшилось, а количество лактобацилл повысилось, что говорит об активации колонизационной резистентности микрофлоры влагалища. У испытателей, у которых до изоляции обнаруживалась существенная обсеменённость анаэробной условно-патогенной микрофлорой, количество всех анаэробов, включая лактобациллы, снижалось. Эубиотический индекс, рассчитанный для цервикального канала, после 3-суточной иммерсии снизился. Полученные данные свидетельствуют о том, что после 3-суточной изоляции наблюдалось ухудшение состояния микрофлоры.
 657-663
657-663


Взаимодействие возбудителей сапронозов с клетками наземного растения воробейника краснокорневого
Аннотация
Введение. Существенную роль в экологии возбудителей сапронозов Yersinia pseudotuberculosis и Listeria monocytogenes и в эпидемиологии вызываемых ими инфекций играют наземные растения, употребляемые в пищу. Эти микроорганизмы часто обнаруживаются на растительных субстратах, они размножаются на разных овощных культурах и корнеплодах. В связи с этим актуально изучение жизнеспособности и биологической активности Y. pseudotuberculosis и L. monocytogenes при контакте с разными наземными растениями, в том числе с теми, которые не употребляются в пищу, но используются для лечения.
Цель исследования — изучение взаимодействия возбудителей сапронозов Y. pseudotuberculosis и L. monocytogenes с каллусными культурами наземного растения воробейника краснокорневого (Lithospermum erythrorhizon Siebold et Zucc).
Материалы и методы. В исследование включены штаммы бактерий Y. pseudotuberculosis 512 1b серотипа, рYV+, 82MD+ и L. monocytogenes NCTC (4b) 10527 из коллекции НИИ эпидемиологии и микробиологии им. Г.П. Сомова, а также культура клеток из корней воробейника краснокорневого, линия ВК-39 (Коллекция ФНЦ биоразнообразия ДВО РАН).
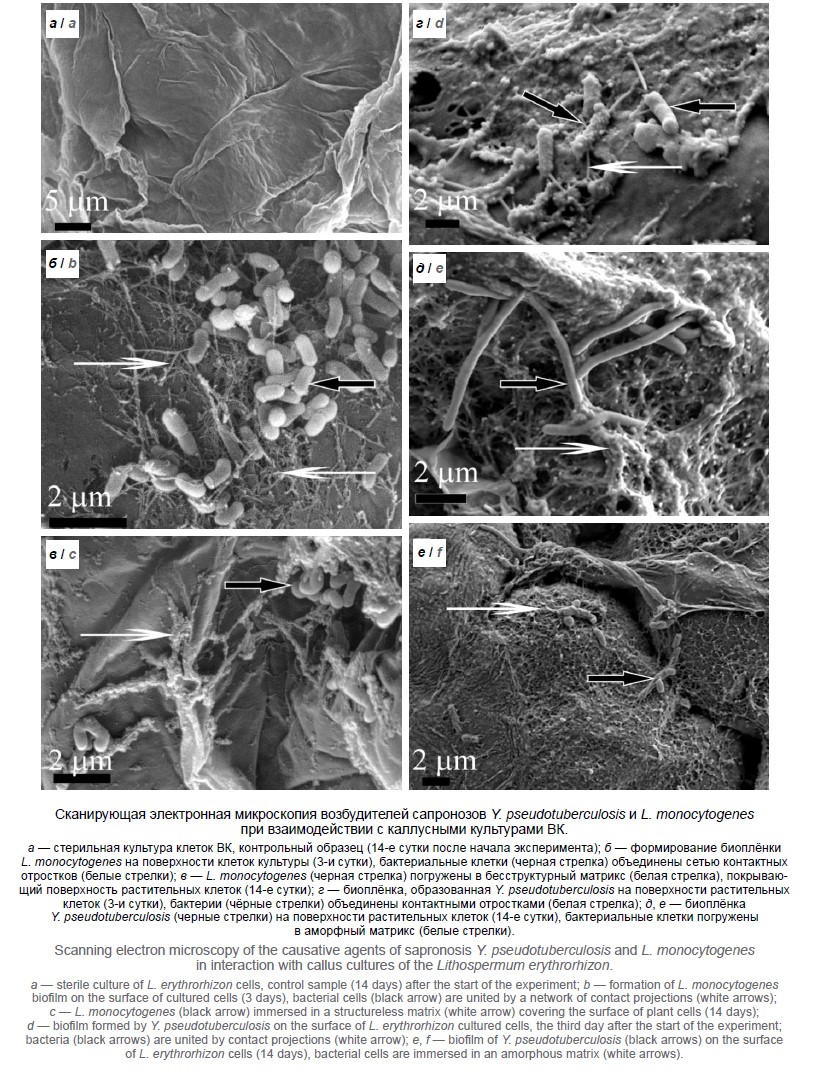
Перед исследованиями Y. pseudotuberculosis и L. monocytogenes культивировали на питательном агаре при рН 7,1–7,2 в течение 18–20 ч. Готовили рабочее разведение микроорганизмов (106 микробных клеток на 1 мл) и наносили их в дозе 100 мкл на поверхность каллусов растений. В динамике через 3 и 14 сут брали пробы материала и готовили их для сканирующей электронной микроскопии.
Результаты. Уже в течение 3–14 сут после начала эксперимента Y. pseudotuberculosis и L. monocytogenes образовывали биоплёнки на поверхности клеток исследуемых растений. При этом впервые обнаружено, что Y. pseudotuberculosis разрушали компоненты оболочки клеток. Заключение. Новые данные, полученные при исследовании, расширяют представления о средах и формах обитания, а также потенциале патогенности возбудителей сапронозов в окружающей среде.
 664-670
664-670


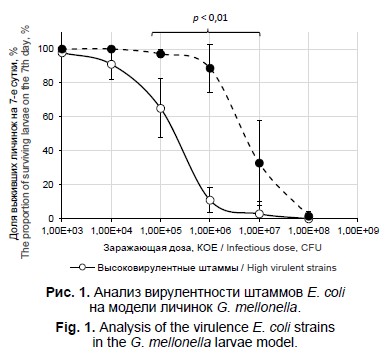
Характеристика вирулентных штаммов Escherichia coli, выделенных от пациентов с урологической инфекцией
Аннотация
Введение. Инфекции мочевыводящих путей (ИМП), вызванные уропатогенными Escherichia coli (UPEC), ежегодно поражают 150 млн человек.
Цель: характеристика внегоспитальных штаммов UPEC, выделенных от пациентов с ИМП в Ярославле в 2016–2017 гг.
Материалы и методы. Чувствительность штаммов UPEC (n = 20) к антимикробным препаратам определяли методом серийных разведений; гены антибиотикорезистентности и вирулентности, филогруппы, О-серогруппы и сиквенс-типы идентифицировали методом ПЦР и полногеномного секвенирования. Вирулентность штаммов изучали на модели личинок Galleria mellonella.
Результаты. Штаммы UPEC отнесены к категориям лекарственно-резистентных (n = 11) и множественно лекарственно-резистентных (n = 9) патогенов. Выявлены гены β-лактамаз blaTEM (n = 10), blaCTX-M (n = 6), интегроны класса 1 (n = 8) и генные кассеты dfrA17-aadA5 (n = 2), dfrA1 (n = 1) и aacA4-cmlA1 (n = 1). Идентифицированы гены вирулентности UPEC: адгезинов fimH, papG, sfaS, focG, afa/draBC, csgA, сидерофоров iroN, fyuA, iutA, факторов противодействия иммунитету макроорганизма ompT и traT, токсинов hlyA, cnf1, usp, транспортёра капсулы kpsMTII, колицина cvaC, островов патогенности I536, II536, III536, IV536, IIJ96 и IICFT073. Определены высоковирулентные и слабовирулентные для личинок G. mellonella штаммы UPEC с LD50 104–105 и 106–107 КО соответственно. Идентифицированы филогруппы A, B1, B2, E и F, серогруппы О2, О4, О6, O9, O11, О15, О18, О25, О75 и O89, известные сиквенс-типы ST14, ST58, ST69, ST73, ST93, ST127, ST131, ST141, ST165, ST297, ST457, ST537, ST744, ST1434 и впервые найденные в данном исследовании ST9239 и ST10102.
Заключение. Выявленное генетическое разнообразие внегоспитальных штаммов UPEC согласуется с мировой тенденцией распространения патогенов человека, обладающих одновременно высокой вирулентностью и множественной лекарственной устойчивостью. Это позволяет проспективно охарактеризовать текущую эпидемиологическую ситуацию, дать прогноз её развития, а также определить оптимальные направления терапии.
 671-684
671-684


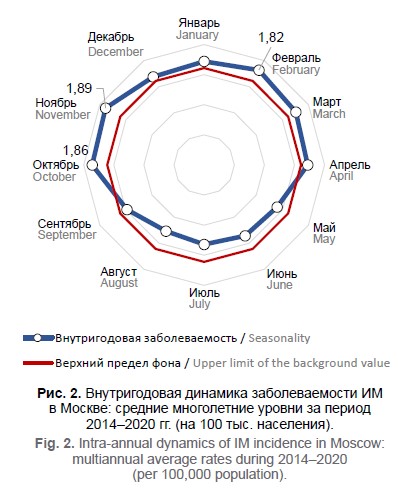
Эпидемиологические особенности инфекции, вызванной вирусом Эпштейна–Барр
Аннотация
Введение. Вирус Эпштейна–Барр (ВЭБ) — один из самых распространённых патогенов - поражённость им населения достигает 90%. В то же время не установлены особенности эпидемического процесса ВЭБ-инфекции. Ранее проведённые исследования посвящены оценке показателей заболеваемости инфекционным мононуклеозом (ИМ) без учёта серологического статуса населения.
Цель работы - выявить эпидемиологические особенности и оценить превалентность серологических маркеров ВЭБ-инфекции для последующей разработки комплекса противоэпидемических мероприятий. Материалы и методы. В Москве анализу подвергнуты данные заболеваемости ИМ (форма № 2 «Сведения об инфекционных и паразитарных заболеваниях») и результаты обследования 138 232 человек на наличие IgG VCA, IgG EBNA, IgM VCA, IgG EA, ДНК ВЭБ в образцах крови и слюны в 2011–2020 гг.
Результаты. Впервые установлены периодичность заболеваемости ИМ с интервалом 9–11 лет и её сильные прямые значимые корреляционные связи с выявлением маркеров активной ВЭБ-инфекции. Для внутригодовой динамики заболеваемости ИМ характерен сезонный подъём в холодный период года с максимальными показателями в октябре, ноябре или феврале, обусловленный выраженным снижением серопревалентности IgG VCA и IgG EBNA. Группами риска по заболеваемости первичной инфекцией являются дети 1–17 лет, что подтверждается достоверно более низкой, по сравнению со взрослыми, частотой выявления маркеров хронической ВЭБ-инфекции (IgG VCA и IgG EBNA) и высокой — IgM VCA и ДНК ВЭБ в крови. Вклад взрослого населения в эпидемический процесс формируется за счёт реактивации хронической инфекции, преимущественно у женщин.
Заключение. Выявленные особенности позволяют дать развёрнутую характеристику эпидемического процесса ВЭБ-инфекции и могут быть использованы для разработки комплекса профилактических и противоэпидемических мероприятий.
 685-696
685-696


Определение чувствительности Mycobacterium tuberculosis к противотуберкулёзным препаратам с помощью полногеномного секвенирования и программного обеспечения «Mykrobe»
Аннотация
Введение. Чувствительность Mycobacterium tuberculosis к противотуберкулёзным препаратам устанавливается с помощью фенотипических и молекулярных методов. Анализ целого генома штаммов M. tuberculosis даёт возможность предсказывать резистентность к лекарствам для большого числа медикаментов. Для этого разработано несколько видов программного обеспечения.
Цель работы - определить чувствительность M. tuberculosis к антитуберкулёзным препаратам с помощью фенотипического и генотипического анализа, а также полногеномного секвенирования с использованием программного обеспечения «Mykrobe».
Материалы и методы. Исследовали 34 мультирезистентных штамма M. tuberculosis, выделенных из клинических материалов 34 пациентов в Болгарии. Все они были подтверждены фенотипически с помощью «BACTEC MGIT 960 System». Для определения резистентности к противотуберкулёзным средствам первого и второго ряда пользовались тестами для линейной гибридизации «Geno Type MTBDR plus v.1.0» и «Geno Type MTBDR sl v.1.0». Штаммы M. tuberculosis секвенировали с помощью «MiSeq». Для электронной резистограммы применяли программное обеспечение «Mykrobe v.0.8.1».
Результаты. Все три метода — фенотипический анализ, генетический анализ и электронная резистограмма с помощью программного обеспечения «Mykrobe» — дали сопоставимые результаты чувствительности/резистентности исследуемых штаммов. Все фенотипически доказанные штаммы, резистентные к рифампицину и изониазиду, были подтверждены на 100% с помощью программного обеспечения «Mykrobe». Мутация С-15Т является маркером для резистентности к изониазиду у исследуемых нами штаммов со сполиготипом SIT41. Мы наблюдали 75% (21/28) совпадения результатов по «BACTEC» и «Mykrobe» в отношении резистентности к этамбутолу. Фенотипически 87% (n = 27) штаммов были устойчивы к стрептомицину, и лишь 59% (n = 19) доказаны программным обеспечением «Mykrobe» как таковые. Сравнивая фенотипическую и генотипическую резистентность к офлоксацину, амикацину и канамицину, мы наблюдали совпадение результатов на 100%.
Выводы. Секвенирование целого генома относительно дорого и трудоёмко, но представляет собой ценный инструмент эпидемиологического генотипирования и определения восприимчивости к лекарственным средствам.
 697-705
697-705


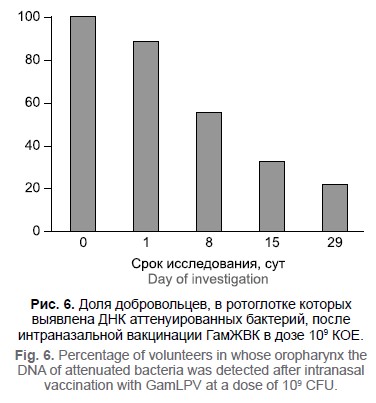
Иммуногенность препарата «Живая вакцина интраназального применения для профилактики коклюша» (ГамЖВК) при однократном применении у здоровых добровольцев
Аннотация
Введение. Значительный рост заболеваемости коклюшем в мире, в том числе среди подростков и взрослых, распространение стёртых форм заболевания, бессимптомное носительство бактерий Bordetella pertussis и обусловленная этим потребность в массовой ревакцинации разных возрастных групп населения обосновывают необходимость разработки новых противококлюшных вакцин. В НИЦЭМ им. Н.Ф. Гамалеи разработана живая коклюшная вакцина интраназального применения для профилактики коклюша (ГамЖВК). Вакцина ГамЖВК прошла доклинические исследования, доказавшие её безопасность и эффективность в экспериментах на мелких лабораторных животных и нечеловекообразных обезьянах. Показана её безопасность в клинических исследованиях на здоровых добровольцах.
Цель исследования - оценка иммуногенности разных доз препарата ГамЖВК при первом применении у здоровых добровольцев.
Материалы и методы. Исследование проводилось как рандомизированное плацебо-контролируемое, слепое, с последовательным включением добровольцев и эскалацией дозы. Идентификатор исследования в базе clinicaltrials.gov: NCT03137927 (A Phase I Clinical Study of a GamLPV, a Live Intranasal Bordetella Pertussis Vaccine). Определены параметры гуморального и клеточного иммунного ответа в динамике: уровни специфических IgM-, IgG- и IgA-антител в сыворотке крови добровольцев и количество цитокинов интерлейкина-17, фактора некроза опухоли-α, интерферона-γ, продуцируемых после специфической индукции in vitro мононуклеарами периферической крови вакцинированных добровольцев. Оценена динамика персистенции аттенуированных бактерий в носоглотке вакцинированных добровольцев.
Результаты. Интраназальная вакцинация добровольцев препаратом ГамЖВК приводит к формированию специфического гуморального (IgG и IgА) и клеточного иммунного ответа. Показан дозозависимый характер продукции иммуноглобулинов и цитокинов. Аттенуированные бактерии длительно персистируют в носо/ротоглотке вакцинированных добровольцев.
Обсуждение. Хорошая переносимость всех тестированных доз препарата обосновывает выбор для дальнейшего исследования дозы вакцины, равной 4 × 109 КОЕ. На следующем этапе будут изучены безопасность и иммуногенность двукратной вакцинации добровольцев.
 706-720
706-720


ОБЗОРЫ
Молекулярные детерминанты резистентности Salmonella enterica к антибиотикам
Аннотация
Нетифоидные штаммы Salmonella enterica представляют большую опасность для здоровья человека. Проблема сальмонеллёзов осложняется прогрессирующим распространением нечувствительности к антибиотикам среди клинических и сельскохозяйственных штаммов S. enterica. Настоящий обзор литературы обобщает современные сведения о механизмах устойчивости S. enterica к антибиотикам и иллюстрирует многообразие и сложность молекулярных систем, обеспечивающих антибиотикорезистентность (АР) у S. enterica. Описан спектр природной резистентности и тщательно охарактеризованы адаптивные (приобретённые) механизмы устойчивости к представителям основных классов антибиотиков, включая β-лактамы, фторхинолоны, аминогликозиды, тетрациклины, нитрофураны, сульфонамиды, фосфомицин, хлорамфеникол (левомицетин) и полимиксины (колистин). Перечислены генетические детерминанты резистентности, передающиеся горизонтальным путём. В обзоре проанализированы только те варианты молекулярных механизмов АР, клиническая значимость которых была доказана комплексом корректных генетических (секвенирование) и биохимических (подтверждение спектра гидролизируемых β-лактамов) исследований. Описаны общие характеристики устойчивости к антибиотикам у нетифоидных сальмонелл. У многих штаммов S. enterica наблюдаются сочетание различных механизмов АР и множественная резистентность. Поднят вопрос о неоднородности распространения резистентности среди различных групп/серотипов внутри вида S. enterica. В частности, некоторые клональные комплексы с признаками резистентности оказываются более успешными патогенами человека и животных. Сальмонеллы, как и большинство других бактерий, демонстрируют неканонический вид устойчивости к антибиотикам — биоплёночную резистентность, которая реализуется за счёт нескольких механизмов, главными из которых являются фильтрующая/сорбционная способность биоплёночного матрикса и трансформация биоплёночных клеток в дормантные и персистирующие формы.
Несмотря на то что функциональная значимость молекулярных ансамблей, определяющих устойчивость к антибиотикам, однотипна для всех энтеробактерий, конкретизация механизмов резистентности у сальмонелл является необходимым звеном для разработки молекулярно-диагностических систем оценки чувствительности сальмонелл к антимикробным препаратам.
 721-730
721-730