COVID-19: эволюция пандемии в России. Сообщение II: динамика циркуляции геновариантов вируса SARS-CoV-2
- Авторы: Акимкин В.Г.1, Попова А.Ю.2, Хафизов К.Ф.1, Дубоделов Д.В.1, Углева С.В.1, Семененко Т.А.3, Плоскирева А.А.1, Горелов А.В.1, Пшеничная Н.Ю.1, Ежлова Е.Б.2, Летюшев А.Н.2, Демина Ю.В.2, Кутырев В.В.4, Максютов Р.А.5, Говорун В.М.6, Дятлов И.А.7, Тотолян А.А.8, Куличенко А.Н.9, Балахонов С.В.10, Рудаков Н.В.11, Троценко О.Е.12, Носков А.К.13, Зайцева Н.Н.14, Топорков А.В.15, Лиознов Д.А.16, Андреева Е.Е.17, Микаилова О.М.18, Комаров А.Г.19, Ананьев В.Ю.20, Молдованов В.В.21, Логунов Д.Ю.3, Гущин В.А.3, Дедков В.Г.8, Черкашина А.С.1, Кузин С.Н.1, Тиванова Е.В.1, Кондрашева Л.Ю.1, Саенко В.В.1, Селезов С.Ю.1, Гасанов Г.А.1, Сванадзе Н.Х.1, Глазов М.Б.1, Остроушко А.А.1, Миронов К.О.1, Есьман А.С.1, Осина Н.А.4, Боднев С.А.5, Комиссаров А.Б.16, Даниленко Д.М.16, Богун А.Г.7, Скрябин Ю.П.7, Лопатовская К.В.10, Штрек С.В.11, Волынкина А.С.9, Гладких А.С.8, Котова В.О.12, Водопьянов А.С.13, Новикова Н.А.14, Сперанская А.С.6, Самойлов А.Е.6, Неверов А.Д.1, Шпак И.М.15
-
Учреждения:
- Центральный научно-исследовательский институт эпидемиологии
- Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека
- Национальный исследовательский центр эпидемиологии и микробиологии имени почётного академика Н.Ф. Гамалеи
- Российский научно-исследовательский противочумный институт «Микроб»
- Государственный научный центр вирусологии и биотехнологии «Вектор»
- Институт дезинфектологии
- Государственный научный центр прикладной микробиологии и биотехнологии
- Санкт-Петербургский научно-исследовательский институт эпидемиологии и микробиологии имени Пастера
- Ставропольский научно-исследовательский противочумный институт
- Иркутский научно-исследовательский противочумный институт Сибири и Дальнего Востока
- Омский научно-исследовательский институт природно-очаговых инфекций
- Хабаровский научно-исследовательский институт эпидемиологии и микробиологии
- Ростовский-на-Дону противочумный институт
- Нижегородский научно-исследовательский институт эпидемиологии и микробиологии имени академика И.Н. Блохиной
- Волгоградский научно-исследовательский противочумный институт
- Научно-исследовательский институт гриппа имени А.А. Смородинцева
- Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека по городу Москве
- Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека по Московской области
- Диагностический центр (Центр лабораторных исследований) Департамента здравоохранения города Москвы
- Федеральный центр гигиены и эпидемиологии
- Центр гигиены и эпидемиологии в городе Москве
- Выпуск: Том 99, № 4 (2022)
- Страницы: 381-396
- Раздел: ОРИГИНАЛЬНЫЕ ИССЛЕДОВАНИЯ
- URL: https://microbiol.crie.ru/jour/article/view/1303
- DOI: https://doi.org/10.36233/0372-9311-295
- ID: 1303
Цитировать
Аннотация
Актуальность. Продолжающаяся пандемия новой коронавирусной инфекции (COVID-19) определяет актуальность проведения молекулярного-генетического мониторинга распространения SARS-CoV-2 среди населения Российской Федерации.
Цель работы — анализ динамики циркуляции геновариантов вируса SARS-CoV-2 на территории России.
Материалы и методы. Проведён анализ динамики циркуляции геновариантов вируса SARS-CoV-2 с 28.12.2020 по 26.06.2022 на территории России. Использованы материалы отчёта Роспотребнадзора № 970 «Информация о случаях инфекционных заболеваний у лиц с подозрением на новую коронавирусную инфекцию», Российской платформы агрегации информации о геномах вирусов (VGARus). Наличие РНК SARS-CoV-2 было подтверждено методом полимеразной цепной реакции в режиме реального времени с обратной транскрипцией. Для проведения амплификации фрагментов генома и последующего секвенирования использовались разработанные в ЦНИИ Эпидемиологии праймерные панели.
Результаты и обсуждение. С помощью российской платформы VGARus, развёрнутой на базе ЦНИИ Эпидемиологии, получены данные о мутационной изменчивости SARS-CoV-2. Мониторинг циркуляции геновариантов SARS-CoV-2 на территории России с 28.12.2020 по 26.06.2022 выявил доминирование геновариантов Delta и Omicron на различных этапах эпидемии.
Заключение. Данные молекулярно-генетических исследований являются важнейшим компонентом эпидемиологического надзора для принятия управленческих решений по предотвращению дальнейшего распространения SARS-CoV-2 и формируют основу для создания новых вакцинных препаратов.
Ключевые слова
Полный текст
ВВЕДЕНИЕ
Интенсивное развитие эпидемического процесса новой инфекции COVID-19 (Corona Virus Disease 2019), этиологически связанной с коронавирусом SARS-CoV-2 (Severe acute respiratory syndrome-related coronavirus 2), в глобальном масштабе создало благоприятные эволюционные условия для появления генетических вариантов возбудителя, которые приобретают новые патогенные свойства. Этому направлению эволюции SARS-CoV-2 способствуют, с одной стороны, преимущества тех вариантов возбудителя, которые активнее передаются от человека к человеку, и, с другой, — ограничение распространения вариантов вируса, которые вызывают более тяжёлое клиническое течение и, соответственно, госпитализацию пациентов, что уменьшает число эффективных контактов. Следует подчеркнуть, что на современном этапе эпидемический процесс COVID-19 в мире находится в состоянии неустойчивого динамического равновесия и даже незначительное увеличение трансмиссивности возбудителя, при прочих равных условиях, способно привести к росту заболеваемости [1–7].
Как и другие РНК-вирусы, SARS-CoV-2, адаптируясь к своим новым хозяевам — людям, подвержен генетической эволюции, что приводит к мутациям в вирусном геноме, которые могут изменять патогенный потенциал вируса. В связи с продолжающимся появлением множества вариантов SARS-CoV-2 Центр по контролю и профилактике заболеваний США и Всемирная организация здравоохранения (ВОЗ) независимо друг от друга создали системы классификации для разделения возникающих мутаций вируса на несколько подгрупп на основании их влияния на трансмиссивность, летальность и ответ на терапию.
Хотя между этими двумя классификациями имеются различия, в некотором приближении к первой группе вариантов, вызывающих озабоченность (variants of concern — VOCs), можно отнести те, которые имеют ряд доказанных признаков, таких как повышенная контагиозность, тяжёлое течение заболевания, увеличение числа летальных исходов и значительное снижение нейтрализации антителами, образовавшимися в ответ на предыдущую инфекцию или вакцинацию. На текущий момент к указанному классу, согласно классификации ВОЗ, принадлежат варианты Delta (B.1.617.2) и Omicron (B.1.1.529)1. Ко второй группе штаммов, представляющих интерес (variants of interest — VOIs), относят варианты со специфическими генетическими маркерами, ассоциированными с изменениями в связывании с рецепторами, ослаблением нейтрализации антителами, увеличением трансмиссивности, снижением эффективности лечения с прогнозируемым нарастанием тяжести заболевания. В третью группу входят варианты под наблюдением (variants under monitoring — VUM), для которых имеются данные, указывающие на потенциальное влияние на скорость передачи вируса и эффективность лечения, но их доля со временем снизилась до практически нулевого уровня. В классификации Центра по контролю и профилактике заболеваний США существует ещё одна категория — варианты с серьёзными последствиями (variant of high consequence — VOHC), для которых присутствуют убедительные доказательства того, что имеющиеся стратегии диагностики, профилактики и лечения гораздо менее эффективны, чем для ранее циркулирующих форм. Однако на сегодняшний день отсутствуют штаммы, обозначенные как VOHC2. В настоящее время во всём мире эти аббревиатуры стали новыми устойчивыми понятиями определения вариантов коронавируса, вызывающего COVID-19.
Чрезвычайно важной является оценка динамики распространения известных и новых геновариантов SARS-CoV-2, циркулирующих на территории России. В соответствии с Постановлением Правительства РФ от 23.03.2021 № 448 «Об утверждении Временного порядка предоставления данных расшифровки генома возбудителя новой коронавирусной инфекции (COVID-19)» в ЦНИИ Эпидемиологии Роспотребнадзора разработана и введена в действие Российская платформа агрегации информации о геномах вирусов (Virus Genome Aggregator of Russia — VGARus)3. База данных VGARus содержит информацию о нуклеотидных последовательностях вирусов SARS-CoV-2 и их мутациях, распространённых в тех или иных регионах России, и может быть использована для хранения, систематизации и выборки данных для выявления мутаций и определения штаммов вирусов.
VGARus дает возможность постоянно вести мониторинг мутационной изменчивости SARS-CoV-2, предоставляя важнейшие данные для обнаружения новых геновариантов и отслеживания их распространённости на территории России. Молекулярно-генетические исследования являются основой для принятия управленческих решений в области профилактических и противоэпидемических мероприятий по предотвращению дальнейшего распространения SARS-CoV-2 и формируют платформу для создания новых вакцинных препаратов [8–10].
Цель — анализ динамики циркуляции геновариантов вируса SARS-CoV-2 на территории России.
МАТЕРИАЛЫ И МЕТОДЫ
Исследование выполнено в ЦНИИ Эпидемиологии Роспотребнадзора. Проведён анализ динамики циркуляции геновариантов вируса SARS-CoV-2 с 28.12.2020 по 24.04.2022 на территории России. Информация о пациентах извлечена из базы данных, сформированной на основе материалов формы отчёта Роспотребнадзора № 970 «Информация о случаях инфекционных заболеваний у лиц с подозрением на новую коронавирусную инфекцию». Указанным пациентам присвоен код МКБ-10 U07.1 «COVID-19, вирус идентифицирован»: COVID-19 подтверждён лабораторными исследованиями, независимо от тяжести клинических признаков или симптомов. В исследовании использовали материалы национальной платформы по агрегации данных о геномах SARS-CoV-2 — VGARus, Централизованной базы данных для построения эпидемиологической аналитики по новой коронавирусной инфекции (COVID-19) и программы для ЭВМ «Эпидемиологическая аналитика по новой коронавирусной инфекции (COVID-19)» [11][12].
Лабораторные исследования проводили в соответствии с МР 3.1.0169-20 «Лабораторная диагностика COVID-19» и другими нормативными документами. Биологическим материалом для исследования являлись мазки из носа, носоглотки и/или горла, промывные воды бронхов, полученные при фибробронхоскопии (бронхоальвеолярный лаваж), (эндо)трахеальный, назофарингеальный аспират, мокрота, биопсийный или аутопсийный материал дыхательных путей.
Наличие РНК SARS-CoV-2 подтверждено методом полимеразной цепной реакции в режиме реального времени с обратной транскрипцией (ОТ-ПЦР) с применением тест-систем АмплиСенс® Cov-Bat-FL (№ РЗН 2014/1987 от 07.04.2020) и на основе LAMP АмплиСенс® SARS-CoV-2 (№ РЗН 2021/13357 от 03.02.2021). Для количественного определения РНК SARS-CoV-2 методом ОТ-ПЦР использован набор реагентов АмплиСенс® COVID19-FL (№ РЗН 2021/14026 от 09.04.2021). Секвенирование в ЦНИИ Эпидемиологии осуществлялось на платформе «Illumina MiSeq» с использованием реагентов MiSeq Reagent Kit v2 (PE 150 + 150 или PE 250 + 250 циклов) или MiSeq Reagent Kit v3 (PE 300 + 300 циклов), а также «Illumina NextSeq 2000» с использованием реагентов NextSeq 1000/2000 P2 (300 циклов) v3. Все последовательности, полученные в исследовании, загружены в базу данных VGARus. Помимо указанных подходов, другими организациями, участвующими в наполнении базы данных VGARus, использовались иные методы амплификации нуклеиновых кислот и технологии высокопроизводительного секвенирования.
Для статистической обработки использованы стандартные методы описательной статистики «Microsoft Excel» и «Statistica v.12.0» («StatSoft»).
РЕЗУЛЬТАТЫ
В рамках Постановления Правительства РФ от 23.03.2021 № 448 образован Консорциум, в состав которого входят научные организации Роспотребнадзора, а также научные организации других ведомств. На сегодняшний день к системе подключена 131 организация, из них 40 выполняют секвенирование [13].
В результате реализации указанного Постановления удалось создать ресурс, уникальный по охвату территории страны и полноте сопроводительной информации, включающей сведения, необходимые для эпидемиологического анализа. Алгоритм работы с данными VGARus позволяет осуществлять оперативный и ретроспективный анализ распространения генетических вариантов SARS-CoV-2 с учётом новейших сведений о генетическом разнообразии возбудителя COVID-19.
В настоящее время проводится регулярный мониторинг эволюции вируса с целью своевременного реагирования на появление его новых потенциально опасных вариантов и корректировки мер для предотвращения распространения COVID-19. Обязательная регистрация проводится в соответствии с Постановлением Правительства РФ № 448 «Об утверждении Временного порядка предоставления данных расшифровки генома возбудителя новой коронавирусной инфекции (COVID-19)».
Специалистами ЦНИИ Эпидемиологии созданы биоинформатические средства анализа данных для выявления мутаций и отображения их принадлежности к эпидемиологически значимым штаммам. Продолжается депонирование данных секвенирования, включая метаданные.
Регистрируемые в базе клинические образцы автоматически получают внутренний регистрационный номер, затем к информации об образце добавляют результат секвенирования нуклеотидной последовательности варианта SARS-CoV-2. Запущенные в базе данных алгоритмы в автоматическом режиме проводят анализ мутаций и идентификацию варианта SARS-CoV-2 в каждом образце. После загрузки нуклеотидной последовательности вируса система автоматически запускает процесс валидации сиквенса, анализ принадлежности к тому или иному геноварианту, а также (в случае, если это полный геном) определяет геновариант по системе Pangolin. Платформа собирает информацию из разных источников: VGARus, эпидемиологические данные Роспотребнадзора, данные с портала «СтопКоронавирус», ВОЗ, демографические и социальные данные по России (статистика, справочники). Помимо этого, в рамках платформы VGARus разработан модуль «Эпидемиологическая аналитика по новой коронавирусной инфекции», который позволяет анализировать в реальном времени данные, загруженные на платформу VGARus.
Динамический мониторинг мутационной изменчивости коронавирусов, выявленных на территории России, осуществляется с декабря 2020 г., при обнаружении первого случая завоза (28.12.2020) геноварианта Alpha (B.1.1.7).
По данным национальной базы VGARus, зарегистрировано 130 355 последовательностей вируса SARS-CoV-2, из них 67 451 (51,7%) полногеномный сиквенс, 62 904 (48,3%) фрагментных сиквенса.
Среди загруженных в базу VGARus последовательностей 112 344 (86,2%) относятся по классификации ВОЗ (согласно пересмотру классификации от 26.11.2021) к вариантам VOCs. Каждый из этих вариантов отличается от Уханьского специфичным набором мутаций, из них к варианту Alpha относятся 1217 (0,9%) последовательностей, Beta — 94 (< 1%), Gamma — 26 (< 1%), Delta — 58 530 (44,9%), Omicron — 52 477 (40,2%). Не относятся к вариантам VOCs 18 011 (13,8%) загруженных последовательностей.
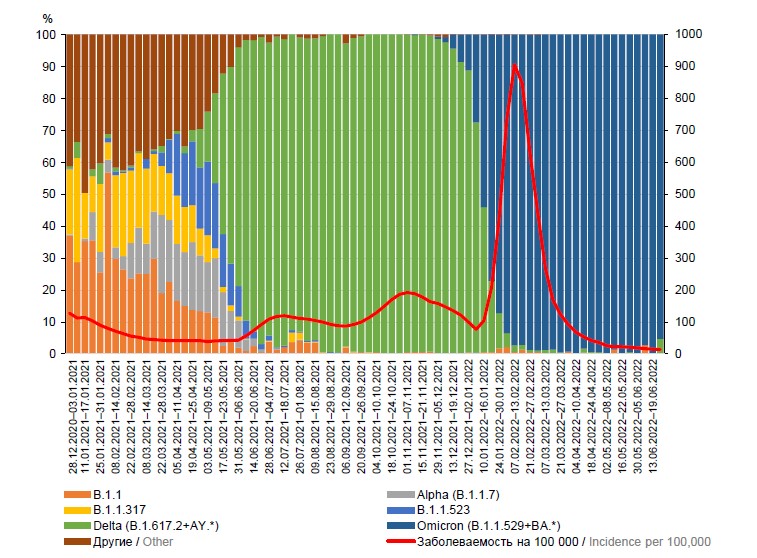
Геновариант Alpha был распространён на территории России зимой 2021 г. Геноварианты Beta и Gamma также встречались в начале 2021 г., однако заметного распространения не получили. Геновариант Delta распространился на территории страны во второй половине апреля 2021 г. и превалировал до января 2022 г. Геновариант Omicron обнаружен в стране в декабре 2021 г. и с января 2022 г. является доминирующим на территории России. Динамика выявленных геновариантов SARS-CoV-2 за 2020–2022 гг. и заболеваемость населения COVID-19 (на 100 тыс. населения) на территории РФ представлены на рис. 1.
В результате полногеномного секвенирования с 30.03.2020 по 26.06.2022 на платформу VGARus загружен 67 451 идентифицированный образец. Из них 724 (1,07%) образца классифицированы как геновариант B.1.1.523, 1329 (1,97%) — B.1.1.317, 3903 (5,79%) — B.1.1, 802 (1,19%) — Alpha (B.1.1.7), 3678 (5,45%) — другие геноварианты, 22818 (33,83%) — Omicron (B.1.1.529+BA.*), 34 197 (50,69%) образцов — Delta (B.1.617.2+AY.*) (рис. 2).
Распределение геновариантов вируса SARSCoV-2 по периодам эпидемического роста заболеваемости COVID-19 на территории России за 2020– 2022 гг., свидетельствующее о доминировании геновариантов Delta (B.1.617.2+AY.*) и Omicron (B.1.1.529+BA.*), отражено в табл. 1.
Таблица 1. Распределение геновариантов вируса SARS-CoV-2 по периодам эпидемического роста заболеваемости COVID-19 на территории России (2020–2022 гг.)
Table 1. Distribution of SARS-CoV-2 genetic variants by periods of the epidemic increase in the COVID-19 incidence in Russia (2020–2022)
Период Period | Число образцов Number of samples | Геноварианты, абс. (%) / Genovariants, abs. (%) | ||||||
B.1.1 | B.1.1.317 | B.1.1.523 | Alpha (B.1.1.7) | Delta (B.1.617.2+AY.*) | Omicron (B.1.1.529+BA.*) | другие other | ||
I период I period 30.03.202030.08.2020 | 1701 | 1056 (62,08) | 55 (3,28) | - | - | - | - | 590 (34,69) |
II период II period 31.08.2020 09.05.2021 | 7417 | 2497 (33,67) | 1202 (16,21) | 378 (5,10) | 536 (7,23) | 121 (1,63) | - | 2683 (36,17) |
III период III period 10.05.202112.09.2021 | 10 602 | 126 (1,19) | 67 (0,63) | 345 (3,25) | 266 (2,51) | 9502 (89,62) | - | 296 (2,79) |
IV период IV period 19.09.202109.01.2022 | 23 315 | 21 (0,09) | 2 (0,009) | 1 (0,004) | - | 21 795 (93,48) | 1420 (6,09) | 76 (0,33) |
V период V period 10.01.202226.06.2022 | 24 416 | 203 (0,83) | 3 (0,012) | - | - | 2779 (11,38) | 21 398 (87,64) | 33 (0,14) |
Всего Total | 67 451 | 3903 (5,79) | 1329 (1,97) | 724 (1,07) | 802 (1,19) | 34 197 (50,69) | 22 818 (33,83) | 3678 (5,45) |
Генетический вариант Delta (B.1.617.2+AY.*) с мая по декабрь 2021 г. превалировал на территории РФ, его доля среди выявленных вариантов составляла до 100%. Доминирующим во все месяцы наблюдения с момента начала регистрации геноварианта Delta являлся вариант, которому с 26.11.2021 классификатор Pangolin присвоил название AY.122 (83,3%).
Помимо AY.122 наиболее часто встречались такие субварианты Delta, как «материнский» B.1.617.2 (9,1%), AY.126 (3,0%) и др. Прочие субварианты представлены единичными образцами и составляли суммарно 4,6% от всех последовательностей Delta (рис. 3).
Всего на территории России выделено 30 сублиний геноварианта Delta, 5% проанализированных полных геномов линии Delta (B.1.617.2+AY.*) представлены на рис. 4.
При анализе динамики субвариантов линии Delta (B.1.617.2+AY.*), выделенных в России, по данным национальной базы VGARus, надо отметить, что в мае 2021 г. линия Delta в основном была представлена сублиниями В.1.617.2 (38,7%), AY.122 (33,8%) и реже встречающимися сублиниями, затем произошла их диссоциация, и до 80% в общей структуре стала занимать сублиния AY.122 (рис. 5).
Вариант Omicron начал стремительное распространение с декабря 2021 г., и в настоящее время он полностью доминирует на территории России (100% всех исследованных образцов). Анализ данных национальной базы VGARus позволил выявить диссоциацию генетической линии Omicron на территории России с наибольшей частотой циркуляции субвариантов BA.1 (54,5%), BA.1.1 (21,6%) и BA.2 (23,8%). Субвариант BA.3 не получил столь значимого распространения и на сегодняшний день составляет менее 0,1% в общей структуре популяции Omicron (рис. 6).
Следует отметить, что в структуре линии Omicron (B.1.1.529+BA.*) с 01.03.2022 начал преобладать субвариант ВА.2, что совпало со снижением уровня заболеваемости COVID-19. В общей структуре на сегодняшний день он занимает свыше 80% (рис. 7).
Таким образом, проанализировав структуру доминирующих геновариантов линии Delta (B.1.617.2+AY.*) и линии Omicron (B.1.1.529+BA.*), можно отметить, что неоднородность и быстрая смена патогенных свойств и конгтагиозности вируса однозначно влияют на течение эпидемического процесса. Доказательством этому утверждению служат показатели динамики проявлений эпидемического процесса и тяжести течения заболевания (табл. 2). Максимальный уровень заболеваемости в 2020 г. (преобладание Уханьского штамма) составил 51,31 (на 100 тыс. населения); максимальный уровень заболеваемости в 2021 г. (преобладание штамма Delta) — 192,45 (на 100 тыс. населения); максимальный уровень заболеваемости в 2022 г. (преобладание штамма Omicron) — 905,37 (на 100 тыс. населения).
Таблица 2. Сравнительная характеристика (динамика) проявлений эпидемического процесса COVID-19 с учётом эволюции возбудителя
Table 2. Comparative analysis (dynamics) of the manifestations of the COVID-19 epidemic process, considering the evolution of the pathogen
Проявления эпидемического процесса Manifestations of the epidemic process | Уханьский геновариант Wuhan genetic variant | Геновариант Delta Delta genetic variant | Геновариант Omicron Omicron genetic variant |
Заболеваемость на 100 тыс. населения Incidence per 100,000 population | 51,31 | 192,45 | 905,37 (рост в 17,6 раза; р < 0,05) 905.37 (a 17.6-fold increase; p < 0.05) |
Удельный вес тяжёлых форм инфекции, % Percentage of severe cases of infection, % | 4,5 | 2,6 | 0,4% (снижение в 11,3 раза; р < 0,05) 0.4% (a 11.3-fold decrease; p < 0.05) |
Удельный вес циркуляции коронавируса среди условно здорового населения, %* Percentage of circulating coronaviruses among relatively healthy population, %* | 10-12 | 13-16 | 30-37% (рост в 3 раза; р < 0,05) 30-37% (a 3-fold increase; p < 0.05) |
Удельный вес детей среди заболевших, % Percentage of children among affected individuals, % | 10 | 12 | 18% (рост в 1,8 раза; р < 0,05) 18% (a 1.8-fold increase; p < 0.05) |
Примечание. *По данным города Москвы и Московской области (n = 2 366 527).
Note. *Based on the data for Moscow and Moscow Region (n = 2,366,527).
ОБСУЖДЕНИЕ
В первый год присутствия SARS-CoV-2 в человеческой популяции в его геноме не наблюдалось нуклеотидных замен, которые бы привели к заметным изменениям свойств патогена. Однако, поскольку сохранение возбудителя как биологического вида невозможно без эволюционного развития, начинается расширение диапазона гетерогенности популяции коронавируса за счёт циркуляции как маловирулентных, так и вирулентных вариантов с последующим стабилизирующим отбором и становлением эпидемического варианта возбудителя. Первые значимые VOCs выявлены в конце 2020 г. — начале 2021 г.: Alpha (B.1.1.7) в Великобритании, Beta (B.1.351) в Южной Африке, Gamma (P.1) в Бразилии и Delta (B.1.617.2) в Индии4. Возникшие мутации изменили аминокислотную последовательность спайкового (S) белка, который после связывания рецептора ACE2 определяет проникновение вируса в чувствительные клетки человеческого организма и является основным фактором патогенеза COVID-19. Подобные мутации вызывают обоснованные опасения, поскольку от них зависит, станет ли вирус более агрессивным.
В ноябре 2021 г. конец осторожному оптимизму экспертов и надеждам на скорое окончание пандемии COVID-19 положило появление нового варианта коронавируса SARS-CоV-2, впервые идентифицированного в Ботсване и Южно Африканской Республике. Новая линия получила обозначение BA.2, основная линия недавно выявленного варианта коронавируса — BA.1, общее название варианта осталось неизменным — B.1.1.529 по классификации PANGO. 26.11.2021 ВОЗ классифицировала мутировавший вирус как VOC и присвоила ему код Omicron (B.1.1.529+ВА.*). По мнению специалистов, SARS-COV-2 эволюционировал и геномные изменения привели к появлению таких характеристик, как способность вызывать интенсивную передачу вируса, изменять клиническую симптоматику заболевания, уклоняться от иммунного ответа, средств диагностики или лекарственных препаратов. Появление множества кластеров COVID-19 на разных континентах может оказать влияние на эпидемиологическую обстановку, привести к возникновению нового источника риска для здоровья населения во всём мире и появлению новой волны заражений. В этой связи всем странам рекомендуется усилить эпидемиологический надзор; активно проводить геномное секвенирование для эффективного отслеживания циркулирующих вариантов SARS-CoV-2; депонировать полные последовательности генома вируса и сопутствующих метаданных в общедоступную базу данных, например GISAID. В связи с глобальным доминированием геноварианта Omicron необходимо изучать его воздействие на тяжесть заболевания, эффективность противоэпидемических мер, иммунный ответ, нейтрализующую активность антител и другие представляющие интерес параметры5.
К настоящему времени накопился значительный массив данных об эволюционных изменениях генома SARS-CoV-2 с учётом тенденций приобретения новых эпидемиологических свойств. За период циркуляции в человеческой популяции геном SARS-CoV-2, приспосабливаясь к новому хозяину, приобрёл определённое количество нуклеотидных замен.
Пандемия COVID-19 в очередной раз подтвердила правильность теории академика В.Д. Белякова, согласно которой основу развития эпидемического процесса составляет фазовое изменение гетерогенности биологических свойств взаимодействующих популяций возбудителя и человека, основанной на обратных отрицательных связях в процессе саморегуляции, при этом важное значение имеют социальные и природные факторы [14][15]. В соответствии с теорией саморегуляции паразитарных систем изменения связаны не только с генетической вариабельностью, но и с другими полидетерминантными характеристиками возбудителя: при появлении новых геновариантов SARS-CoV-2 стал менее патогенным для человека, но более контагиозным. Это обстоятельство является важным не только для теоретической, но и для практической эпидемиологии, т.к. даёт возможность прогнозировать направления развития эпидемической ситуации.
ЗАКЛЮЧЕНИЕ
Полногеномное секвенирование геновариантов SARS-CoV-2 на территории России с 28.12.2020 по 26.06.2022, проведённое на основе базы данных платформы VGARus, выявило превалирование геновариантов Delta и Omicron и позволило установить, что генетический вариант Delta (B.1.617.2 + AY.*) с мая по декабрь 2021 г. являлся доминирующим на территории страны с преобладанием субварианта AY.122 (83,3%). Вариант Omicron начал стремительное распространение с декабря 2021 г. с диссоциацией генетической линии Omicron и преобладанием субвариантов BA.1, BA.1.1 и BA.2. Сублиния BA.3 не получила столь значимого распространения и на сегодняшний день занимает долю менее 0,1% в общей структуре популяции Omicron. Доля сублинии BA.2 постепенно возрастает (до 80% в структуре сублиний Omicron, выделяемых на 16–17-й неделе 2022 г.). В России зарегистрированы единичные геноварианты ВА.4 и ВА.5, которые не получили эпидемического распространения, а клинические проявления проходят в бессимптомной форме или имеют лёгкое течение заболевания в форме острой респираторной вирусной инфекции.
Установлено, что при появлении новых геновариантов вирус SARS-CoV-2 стал менее патогенным для человека, но более контагиозным. Доказательством этого утверждения служат показатели динамики проявлений эпидемического процесса и тяжести течения заболевания.
Вирус SARS-CoV-2 находится в процессе эволюционного развития, что требует непрерывных научных исследований с использованием передовых методов полногеномного анализа его генетических последовательностей.
Таким образом, в настоящее время молекулярно-генетический мониторинг циркуляции возбудителя SARS-CoV-2 является ведущим направлением эпидемиологического надзора за COVID-19, позволяющим принимать решения по разработке и осуществлению противоэпидемических мероприятий.
Коллектив авторов выражает благодарность организациям, участвовавшим в секвенировании:
- Национальный исследовательский центр эпидемиологии и микробиологии имени почётного академика Н.Ф. Гамалеи, Москва, Россия;
- Российский национальный исследовательский медицинский институт им. Н.И. Пирогова, Москва, Россия;
- Научно-исследовательский институт вакцин и сывороток им. И.И. Мечникова, Москва, Россия;
- Научный медицинский исследовательский центр гематологии, Москва, Россия;
- Федеральный исследовательский центр фундаментальной и трансляционной медицины, Новосибирск, Россия;
- Научный медицинский исследовательский центр акушерства, гинекологии и перинатологии им. В.И. Кулакова, Москва, Россия;
- Тюменский научно-исследовательский институт краевой инфекционной патологии, Тюмень, Россия;
- Центр гигиены и эпидемиологии, Пермь, Россия;
- Федеральный исследовательский центр вирусологии и микробиологии, пос. Вольгинский, Россия;
- Федеральный научный центр исследований и разработки иммунобиологических препаратов им. М.П. Чумакова, Москва, Россия;
- Центр стратегического планирования и управления медико-биологическими рисками здоровью, Москва, Россия.
1. ВОЗ. Отслеживание вариантов вируса SARS-CoV-2; 2021. URL: https://www.who.int/ru/activities/tracking-SARS-CoV-2-variants/tracking-SARS-CoV-2-variants
2. CDC. SARS-CoV-2 Variant Classifications and Definitions; 2022. URL: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html
3. Проект VGARus (Virus Genome Aggregator of Russia). URL: https://genome.crie.ru/app/index
4. WHO. Weekly epidemiological update on COVID-19 — 22 March 2022. URL: https://www.who.int/publications/m/item/weeklyepidemiological-update-on-covid-19---22-march-2022
5. WHO. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern; 2021. URL: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov2-variant-of-concern
Об авторах
Василий Геннадьевич Акимкин
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0003-4228-9044
д.м.н., профессор, академик РАН, директор
Россия, МоскваАнна Юрьевна Попова
Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека
Email: uglevas@bk.ru
ORCID iD: 0000-0002-4315-5307
д.м.н., профессор, Главный государственный санитарный врач Российской Федерации
Россия, МоскваКамиль Фаридович Хафизов
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-5524-0296
PhD, зав. лаб. геномных исследований
Россия, МоскваДмитрий Васильевич Дубоделов
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0003-3093-5731
к.м.н., с.н.с. лаб. вирусных гепатитов отдела молекулярной диагностики и эпидемиологии
Россия, МоскваСветлана Викторовна Углева
Центральный научно-исследовательский институт эпидемиологии
Автор, ответственный за переписку.
Email: uglevas@bk.ru
ORCID iD: 0000-0002-1322-0155
д.м.н., доцент, консультант организационно-методического отдела административно-управленческого подразделения
Россия, МоскваТатьяна Анатольевна Семененко
Национальный исследовательский центр эпидемиологии и микробиологии имени почётного академика Н.Ф. Гамалеи
Email: uglevas@bk.ru
ORCID iD: 0000-0002-6686-9011
д.м.н., профессор, рук. отдела эпидемиологии
Россия, МоскваАнтонина Александровна Плоскирева
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-3612-1889
д.м.н., зам. директора
Россия, МоскваАлександр Васильевич Горелов
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
д.м.н., профессор, член-корреспондент РАН, заместитель директора по научной работе
Россия, МоскваНаталья Юрьевна Пшеничная
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0003-2570-711X
д.м.н., зам. директора
Россия, МоскваЕлена Борисовна Ежлова
Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека
Email: uglevas@bk.ru
ORCID iD: 0000-0002-8701-280X
к.м.н., зам. рук.
Россия, МоскваАлександр Николаевич Летюшев
Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека
Email: uglevas@bk.ru
ORCID iD: 0000-0002-4185-9829
к.м.н., начальник Управления научно-аналитического обеспечения и международной деятельности
Россия, МоскваЮлия Викторовна Демина
Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека
Email: uglevas@bk.ru
ORCID iD: 0000-0003-0538-1992
д.м.н., начальник Управления эпидемиологического надзора
Россия, МоскваВладимир Викторович Кутырев
Российский научно-исследовательский противочумный институт «Микроб»
Email: uglevas@bk.ru
ORCID iD: 0000-0003-3788-3452
д.м.н., профессор, академик РАН, директор
Россия, СаратовРинат Амирович Максютов
Государственный научный центр вирусологии и биотехнологии «Вектор»
Email: uglevas@bk.ru
ORCID iD: 0000-0003-1314-281X
д.б.н., директор
Россия, КольцовоВадим Маркович Говорун
Институт дезинфектологии
Email: uglevas@bk.ru
ORCID iD: 0000-0003-0837-8764
д.б.н., профессор, академик РАН, директор
Россия, МоскваИван Алексеевич Дятлов
Государственный научный центр прикладной микробиологии и биотехнологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-3436-0368
д.м.н., профессор, академик РАН, директор
Россия, ОболенскАрег Артемович Тотолян
Санкт-Петербургский научно-исследовательский институт эпидемиологии и микробиологии имени Пастера
Email: uglevas@bk.ru
ORCID iD: 0000-0003-4571-8799
д.м.н., профессор, академик РАН, зав. лаб. молекулярной иммунологии, директор
Россия, Санкт-ПетербургАлександр Николаевич Куличенко
Ставропольский научно-исследовательский противочумный институт
Email: uglevas@bk.ru
ORCID iD: 0000-0002-9362-3949
д.м.н., профессор, член-корреспондент РАН, директор
Россия, СтавропольСергей Владимирович Балахонов
Иркутский научно-исследовательский противочумный институт Сибири и Дальнего Востока
Email: uglevas@bk.ru
ORCID iD: 0000-0003-4201-5828
д.м.н., профессор, директор
Россия, ИркутскНиколай Викторович Рудаков
Омский научно-исследовательский институт природно-очаговых инфекций
Email: uglevas@bk.ru
ORCID iD: 0000-0001-9566-9214
д.м.н., профессор, директор
Россия, ОмскОльга Евгеньевна Троценко
Хабаровский научно-исследовательский институт эпидемиологии и микробиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0003-3050-4472
д.м.н., профессор, директор
Россия, ХабаровскАлексей Кимович Носков
Ростовский-на-Дону противочумный институт
Email: uglevas@bk.ru
ORCID iD: 0000-0003-0550-2221
к.м.н., директор
Россия, Ростов-на-ДонуНаталья Николаевна Зайцева
Нижегородский научно-исследовательский институт эпидемиологии и микробиологии имени академика И.Н. Блохиной
Email: uglevas@bk.ru
ORCID iD: 0000-0001-5370-4026
д.м.н., директор
Россия, Нижний НовгородАндрей Владимирович Топорков
Волгоградский научно-исследовательский противочумный институт
Email: uglevas@bk.ru
ORCID iD: 0000-0002-3449-4657
д.м.н., доцент, директор
Россия, ВолгоградДмитрий Анатольевич Лиознов
Научно-исследовательский институт гриппа имени А.А. Смородинцева
Email: uglevas@bk.ru
ORCID iD: 0000-0003-3643-7354
д.м.н., директор
Россия, Санкт-ПетербургЕлена Евгеньевна Андреева
Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека по городу Москве
Email: uglevas@bk.ru
ORCID iD: 0000-0001-6687-7276
д.м.н., профессор, рук., главный государственный санитарный врач по городу Москве
Россия, МоскваОльга Михайловна Микаилова
Федеральная служба по надзору в сфере защиты прав потребителей и благополучия человека по Московской области
Email: uglevas@bk.ru
ORCID iD: 0000-0003-3842-6368
к.м.н., Главный государственный санитарный врач по Московской области, рук.
Россия, МоскваАндрей Григорьевич Комаров
Диагностический центр (Центр лабораторных исследований) Департамента здравоохранения города Москвы
Email: uglevas@bk.ru
директор
Россия, МоскваВасилий Юрьевич Ананьев
Федеральный центр гигиены и эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-1670-6791
к.м.н., главный врач
Россия, МоскваВладимир Валерьевич Молдованов
Центр гигиены и эпидемиологии в городе Москве
Email: uglevas@bk.ru
ORCID iD: 0000-0002-5606-4906
д.м.н., главный врач
Россия, МоскваДенис Юрьевич Логунов
Национальный исследовательский центр эпидемиологии и микробиологии имени почётного академика Н.Ф. Гамалеи
Email: uglevas@bk.ru
ORCID iD: 0000-0003-4035-6581
д.б.н., член-корр. РАН, зам. директора по научной работе
Россия, МоскваВладимир Алексеевич Гущин
Национальный исследовательский центр эпидемиологии и микробиологии имени почётного академика Н.Ф. Гамалеи
Email: uglevas@bk.ru
ORCID iD: 0000-0002-9397-3762
к.б.н., зав. лаб. механизмов популяционной изменчивости патогенных микроорганизмов
Россия, МоскваВладимир Георгиевич Дедков
Санкт-Петербургский научно-исследовательский институт эпидемиологии и микробиологии имени Пастера
Email: uglevas@bk.ru
ORCID iD: 0000-0002-5500-0169
к.м.н., зам. директора по научной работе
Россия, Санкт-ПетербургАнна Сергеевна Черкашина
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-7970-7495
к.х.н., рук. научной группы генной инженерии и биотехнологии отдела молекулярной диагностики и эпидемиологии
Россия, МоскваСтанислав Николаевич Кузин
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-0616-9777
д.м.н., профессор, зав. лаб. вирусных гепатитов отдела молекулярной диагностики и эпидемиологии
Россия, МоскваЕлена Валерьевна Тиванова
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0003-1286-2612
рук. направления лабораторной медицины и продвижения лабораторных услуг отдела молекулярной диагностики и эпидемиологии
Россия, МоскваЛариса Юрьевна Кондрашева
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-0147-4262
зав. лаб. полимеразной цепной реакции
Россия, МоскваВалерия Владимировна Саенко
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0003-0952-0830
руководитель научной группы геномных технологий
Россия, МоскваСемен Юрьевич Селезов
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-9451-4341
биоинформатик, лаборатория геномных исследований
Россия, МоскваГасан Алиевич Гасанов
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-0121-521X
аспирант
Россия, МоскваНино Хвичаевна Сванадзе
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-7524-3080
Сванадзе Нино Хвичаевна — врач-эпидемиолог лаб. вирусных
гепатитов отдела молекулярной диагностики и эпидемиологии
ЦНИИ Эпидемиологии
Москва
Россия, МоскваМаксим Борисович Глазов
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-2195-1580
рук. центра по развитию информационных технологий и систем
Россия, МоскваАлексей Александрович Остроушко
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0003-0803-5630
рук. Информационно-аналитической службы
Россия, МоскваКонстантин Олегович Миронов
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-8207-9215
д.м.н., рук. лаб. молекулярных методов изучения генетических полиморфизмов
Россия, МоскваАнна Сергеевна Есьман
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-5456-7649
н.с. лаб. молекулярных методов изучения генетических полиморфизмов
Россия, МоскваНаталия Александровна Осина
Российский научно-исследовательский противочумный институт «Микроб»
Email: uglevas@bk.ru
ORCID iD: 0000-0003-0954-5683
к.б.н., зав. лаб. молекулярной диагностики
Россия, СаратовСергей Александрович Боднев
Государственный научный центр вирусологии и биотехнологии «Вектор»
Email: uglevas@bk.ru
ORCID iD: 0000-0003-0599-3817
к.м.н., в.н.с.
Россия, КольцовоАндрей Борисович Комиссаров
Научно-исследовательский институт гриппа имени А.А. Смородинцева
Email: uglevas@bk.ru
ORCID iD: 0000-0003-1733-1255
зав. лаб. молекулярной вирусологии
Россия, Санкт-ПетербургДарья Михайловна Даниленко
Научно-исследовательский институт гриппа имени А.А. Смородинцева
Email: uglevas@bk.ru
ORCID iD: 0000-0001-6174-0836
к.б.н., зам. директора по научной работе
Россия, Санкт-ПетербургАлександр Геннадьевич Богун
Государственный научный центр прикладной микробиологии и биотехнологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-5454-2495
к.б.н., в.н.с. отд. коллекционных культур
Россия, ОболенскЮрий Павлович Скрябин
Государственный научный центр прикладной микробиологии и биотехнологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-5748-995X
к.б.н., н.с. лаб. антимикробных препаратов
Россия, ОболенскКристина Викторовна Лопатовская
Иркутский научно-исследовательский противочумный институт Сибири и Дальнего Востока
Email: uglevas@bk.ru
ORCID iD: 0000-0002-8772-5842
н.с. лаб. природно-очаговых вирусных инфекций
Россия, ИркутскСергей Владимирович Штрек
Омский научно-исследовательский институт природно-очаговых инфекций
Email: uglevas@bk.ru
ORCID iD: 0000-0002-4509-1212
к.м.н., с.н.с. лаб. зоонозных инфекций
Россия, ОмскАнна Сергеевна Волынкина
Ставропольский научно-исследовательский противочумный институт
Email: uglevas@bk.ru
ORCID iD: 0000-0001-5554-5882
к.б.н., зав. лаб. диагностики вирусных инфекций
Россия, СтавропольАнна Сергеевна Гладких
Санкт-Петербургский научно-исследовательский институт эпидемиологии и микробиологии имени Пастера
Email: uglevas@bk.ru
ORCID iD: 0000-0001-6759-1907
к.б.н., с.н.с. группы молекулярной генетики патогенных микроорганизмов отдела эпидемиологии
Россия, Санкт-ПетербургВалерия Олеговна Котова
Хабаровский научно-исследовательский институт эпидемиологии и микробиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-9824-7025
с.н.с., зав. лаб. эпидемиологии и профилактики вирусных гепатитов и СПИДа
Россия, ХабаровскАлексей Сергеевич Водопьянов
Ростовский-на-Дону противочумный институт
Email: uglevas@bk.ru
ORCID iD: 0000-0002-9056-3231
к.м.н., и.о. зав. группой вирусологии
Россия, Ростов-на-ДонуНадежда Алексеевна Новикова
Нижегородский научно-исследовательский институт эпидемиологии и микробиологии имени академика И.Н. Блохиной
Email: uglevas@bk.ru
ORCID iD: 0000-0002-3710-6648
д.б.н., профессор, зав. лаб. молекулярной эпидемиологии вирусных инфекций
Россия, Нижний НовгородАнна Сергеевна Сперанская
Институт дезинфектологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-6326-1249
к.б.н., с.н.с. Центра геномики и масс-спектрометрии
Россия, МоскваАндрей Евгеньевич Самойлов
Институт дезинфектологии
Email: uglevas@bk.ru
ORCID iD: 0000-0001-8284-3164
к.б.н., с.н.с. Центра геномики и масс-спектрометрии
Россия, МоскваАлексей Дмитриевич Неверов
Центральный научно-исследовательский институт эпидемиологии
Email: uglevas@bk.ru
ORCID iD: 0000-0002-3594-1682
к.б.н., рук. группы биоинформатики
Россия, МоскваИван Михайлович Шпак
Волгоградский научно-исследовательский противочумный институт
Email: uglevas@bk.ru
ORCID iD: 0000-0001-6446-0274
к.м.н., н.с. сектора биоинформационного анализа
Россия, ВолгоградСписок литературы
- Акимкин В.Г., Кузин С.Н., Семененко Т.А., Плоскирева А.А., Дубоделов Д.В., Тиванова Е.В. и др. Характеристика эпидемиологической ситуации по COVID-19 в Российской Федерации в 2020 г. Вестник Российской академии медицинских наук. 2021; 76(4): 412-22. https://doi.org/10.15690/vramn1505
- Пшеничная Н.Ю., Лизинфельд И.А., Журавлев Г.Ю., Плоскирева А.А., Акимкин В.Г. Эпидемический процесс COVID-19 в Российской Федерации: промежуточные итоги. Сообщение 1. Инфекционные болезни. 2020; 18(3): 7-14. https://doi.org/10.20953/1729-9225-2020-3-7-14
- Пшеничная Н.Ю., Лизинфельд И.А., Журавлев Г.Ю., Плоскирева А.А., Еровиченков А.А., Акимкин В.Г. Эпидемический процесс COVID-19 в Российский Федерации: промежуточные итоги. Сообщение 2. Инфекционные болезни. 2021; 19(1): 10-5. https://doi.org/10.20953/1729-9225-2021-1-10-15
- Кутырев В.В., Попова А.Ю., Смоленский В.Ю., Ежлова Е.Б., Демина Ю.В., Сафронов В.А. и др. Эпидемиологические особенности новой коронавирусной инфекции (COVID-19). Сообщение 1: Модели реализации профилактических и противоэпидемических мероприятий. Проблемы особо опасных инфекций. 2020; (1): 6-13. https://doi.org/10.21055/0370-1069-2020-1-6-13
- Краснов Я.М., Попова А.Ю., Сафронов В.А., Федоров А.В., Баданин Д.В., Щербакова С.А. и др. Анализ геномного разнообразия SARS-CoV-2 и эпидемиологических признаков адаптации возбудителя COVID-19 к человеческой популяции (Сообщение 1). Проблемы особо опасных инфекций. 2020; (3): 70-82. https://doi.org/10.21055/0370-1069-2020-3-70-82
- Акимкин В.Г., Кузин С.Н., Семененко Т.А., Шипулина О.Ю., Яцышина С.Б., Тиванова Е.В. и др. Закономерности эпидемического распространения SARS-CoV-2 в условиях мегаполиса. Вопросы вирусологии. 2020; 65(4): 203-11. https://doi.org/10.36233/0507-4088-2020-65-4-203-211
- Акимкин В.Г., Кузин С.Н., Семененко Т.А., Плоскирева А.А., Дубоделов Д.В., Тиванова Е.В. и др. Гендерно-возрастная характеристика пациентов с COVID-19 на разных этапах эпидемии в Москве. Проблемы особо опасных инфекций. 2020; (3): 27-35. https://doi.org/10.21055/0370-1069-2020-3-27-35
- Щелканов М.Ю., Попова А.Ю., Дедков В.Г., Акимкин В.Г., Малеев В.В. История изучения и современная классификация коронавирусов (Nidovirales: Coronaviridae). Инфекция и иммунитет. 2020; 10(2): 221-46. https://doi.org/10.15789/2220-7619-HOI-1412
- Kaptelova V.V., Bukharina A.Y., Shipulina O.Y., Korneenko E.V., Saenko S.S., Lukyanov A.V., et al. Case report: change of dominant strain during dual SARS-CoV-2 infection. BMC Infect. Dis. 2021; 21(1): 959. https://doi.org/10.1186/s12879-021-06664-w
- Борисова Н.И., Котов И.А., Колесников А.А., КаптеловаВ.В., Сперанская А.С., Кондрашева Л.Ю. и др. Мониторинг распространения вариантов SARS-CoV-2 (Coronaviridae: Coronavirinae: Betacoronavirus; Sarbecovirus) на территории Московского региона с помощью таргетного высокопроизводительного секвенирования. Вопросы вирусологии. 2021; 66(4): 269-78. https://doi.org/10.36233/0507-4088-72
- Дубоделов Д.В., Савельер Е.В., Плоскирева А.А., Акимкин В.Г., Глазов М.Б., Гасанов Г.А. и др. Централизованная база данных для построения эпидемиологической аналитики по новой коронавирусной инфекции (COVID-19). Патент РФ № 2021622334; 2021.
- Дубоделов Д.В., Савельер Е.В., Плоскирева А.А., Акимкин В.Г., Глазов М.Б., Гасанов Г.А. и др. Эпидемиологическая аналитика по новой коронавирусной инфекции (COVID-19). Патент РФ № 2021667476; 2021.
- Попова А.Ю., ред. COVID-19: научно-практические аспекты борьбы с пандемией в Российской Федерации. Саратов: Амирит; 2021.
- Беляков В.Д. Внутренняя регуляция эпидемического процесса (ответы на замечания и вопросы, поднятые при обсуждении теории). Журнал микробиологии, эпидемиологии и иммунобиологии. 1987; 64(10): 78-89.
- Беляков В.Д., Голубев Д.Б., Каминский Г.Д., Тец В.В. Саморегуляция паразитарных систем. Ленинград: Медицина; 1987.
Дополнительные файлы